Metabolism-associated molecular classification of hepatocellular carcinoma
- PMID: 31955511
- PMCID: PMC7138397
- DOI: 10.1002/1878-0261.12639
Metabolism-associated molecular classification of hepatocellular carcinoma
Abstract
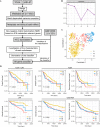
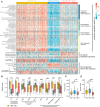
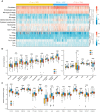
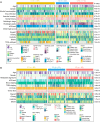
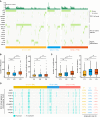
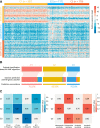
Hepatocellular carcinoma (HCC) is a disease with unique management complexity because it displays high heterogeneity of molecular phenotypes. We herein aimed to characterize the molecular features of HCC by the development of a classification system that was based on the gene expression profile of metabolic genes. Integrative analysis was performed with a metadata set featuring 371 and 231 HCC human samples from the Cancer Genome Atlas and the International Cancer Genome Consortium, respectively. All samples were linked with clinical information. RNA sequencing data of 2752 previously characterized metabolism-related genes were used for non-negative matrix factorization clustering, and three subclasses of HCC (C1, C2, and C3) were identified. We then analyzed the metadata set for metabolic signatures, prognostic value, transcriptome features, immune infiltration, clinical characteristics, and drug sensitivity of subclasses, and compared the resulting subclasses with previously published classifications. Subclass C1 displayed high metabolic activity, low α-fetoprotein (AFP) expression, and good prognosis. Subclass C2 was associated with low metabolic activities and displayed high expression of immune checkpoint genes, demonstrating drug sensitivity toward cytotoxic T-lymphocyte-associated protein-4 inhibitors and the receptor tyrosine kinase inhibitor cabozantinib. Subclass C3 displayed intermediate metabolic activity, high AFP expression level, and bad prognosis. Finally, a 90-gene classifier was generated to enable HCC classification. This study establishes a new HCC classification based on the gene expression profiles of metabolic genes, thereby furthering the understanding of the genetic diversity of human HCC.
Keywords: classification; hepatocellular carcinoma; immune signatures; metabolic genes.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, Herault A, Saric J, Belghiti J, Franco D et al (2007) Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology (Baltimore, MD) 45, 42–52. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

