Ultra-High Mass Resolving Power, Mass Accuracy, and Dynamic Range MALDI Mass Spectrometry Imaging by 21-T FT-ICR MS
- PMID: 31955581
- PMCID: PMC7031845
- DOI: 10.1021/acs.analchem.9b04768
Ultra-High Mass Resolving Power, Mass Accuracy, and Dynamic Range MALDI Mass Spectrometry Imaging by 21-T FT-ICR MS
Abstract
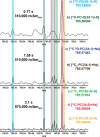
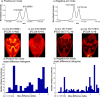
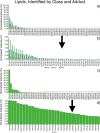
Detailed characterization of complex biological surfaces by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) requires instrumentation that is capable of high mass resolving power, mass accuracy, and dynamic range. Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) offers the highest mass spectral performance for MALDI MSI experiments, and often reveals molecular features that are unresolved on lower performance instrumentation. Higher magnetic field strength improves all performance characteristics of FT-ICR; mass resolving power improves linearly, while mass accuracy and dynamic range improve quadratically with magnetic field strength. Here, MALDI MSI at 21T is demonstrated for the first time: mass resolving power in excess of 1 600 000 (at m/z 400), root-mean-square mass measurement accuracy below 100 ppb, and dynamic range per pixel over 500:1 were obtained from the direct analysis of biological tissue sections. Molecular features with m/z differences as small as 1.79 mDa were resolved and identified with high mass accuracy. These features allow for the separation and identification of lipids to the underlying structures of tissues. The unique molecular detail, accuracy, sensitivity, and dynamic range combined in a 21T MALDI FT-ICR MSI experiment enable researchers to visualize molecular structures in complex tissues that have remained hidden until now. The instrument described allows for future innovative, such as high-end studies to unravel the complexity of biological, geological, and engineered organic material surfaces with an unsurpassed detail.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Lazar A. N.; Bich C.; Panchal M.; Desbenoit N.; Petit V. W.; Touboul D.; Dauphinot L.; Marquer C.; Laprevote O.; Brunelle A.; Duyckaerts C. Time-of-flight secondary ion mass spectrometry (TOF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of Alzheimer disease patients. Acta Neuropathol. 2013, 125 (1), 133–44. 10.1007/s00401-012-1041-1. - DOI - PubMed
-
- Chen Y.; Allegood J.; Liu Y.; Wang E.; Cachon-Gonzalez B.; Cox T. M.; Merrill A. H. Jr.; Sullards M. C. Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal. Chem. 2008, 80 (8), 2780–8. 10.1021/ac702350g. - DOI - PubMed
-
- Scott A. J.; Post J. M.; Lerner R.; Ellis S. R.; Lieberman J.; Shirey K. A.; Heeren R. M. A.; Bindila L.; Ernst R. K. Host-based lipid inflammation drives pathogenesis in Francisella infection. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (47), 12596–12601. 10.1073/pnas.1712887114. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

