Comparative Genomics of Streptococcus thermophilus Support Important Traits Concerning the Evolution, Biology and Technological Properties of the Species
- PMID: 31956321
- PMCID: PMC6951406
- DOI: 10.3389/fmicb.2019.02916
Comparative Genomics of Streptococcus thermophilus Support Important Traits Concerning the Evolution, Biology and Technological Properties of the Species
Abstract
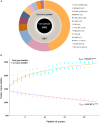
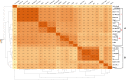
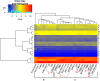
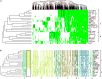
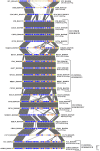
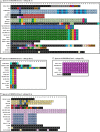
Streptococcus thermophilus is a major starter for the dairy industry with great economic importance. In this study we analyzed 23 fully sequenced genomes of S. thermophilus to highlight novel aspects of the evolution, biology and technological properties of this species. Pan/core genome analysis revealed that the species has an important number of conserved genes and that the pan genome is probably going to be closed soon. According to whole genome phylogeny and average nucleotide identity (ANI) analysis, most S. thermophilus strains were grouped in two major clusters (i.e., clusters A and B). More specifically, cluster A includes strains with chromosomes above 1.83 Mbp, while cluster B includes chromosomes below this threshold. This observation suggests that strains belonging to the two clusters may be differentiated by gene gain or gene loss events. Furthermore, certain strains of cluster A could be further subdivided in subgroups, i.e., subgroup I (ASCC 1275, DGCC 7710, KLDS SM, MN-BM-A02, and ND07), II (MN-BM-A01 and MN-ZLW-002), III (LMD-9 and SMQ-301), and IV (APC151 and ND03). In cluster B certain strains formed one distinct subgroup, i.e., subgroup I (CNRZ1066, CS8, EPS, and S9). Clusters and subgroups observed for S. thermophilus indicate the existence of lineages within the species, an observation which was further supported to a variable degree by the distribution and/or the architecture of several genomic traits. These would include exopolysaccharide (EPS) gene clusters, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs)-CRISPR associated (Cas) systems, as well as restriction-modification (R-M) systems and genomic islands (GIs). Of note, the histidine biosynthetic cluster was found present in all cluster A strains (plus strain NCTC12958T) but was absent from all strains in cluster B. Other loci related to lactose/galactose catabolism and urea metabolism, aminopeptidases, the majority of amino acid and peptide transporters, as well as amino acid biosynthetic pathways were found to be conserved in all strains suggesting their central role for the species. Our study highlights the necessity of sequencing and analyzing more S. thermophilus complete genomes to further elucidate important aspects of strain diversity within this starter culture that may be related to its application in the dairy industry.
Keywords: CRISPR; cheese; genomic islands; horizontal gene transfer; lineage; milk; pan genome; yogurt.
Copyright © 2019 Alexandraki, Kazou, Blom, Pot, Papadimitriou and Tsakalidou.
Figures


References
-
- Anbukkarasi K., Nanda D. K., Umamaheswari T., Hemalatha T., Singh P., Singh R. (2014). Assessment of expression of Leloir pathway genes in wild-type galactose-fermenting Streptococcus thermophilus by real-time PCR. Eur. Food Res. Technol. 239 895–903. 10.1007/s00217-014-2286-9 - DOI
-
- Anbukkarasi K., Umamaheswari T., Hemalatha T., Nanda D. K., Singh P., Rashmi H. M., et al. (2013). Production of low browning Mozzarella cheese: screening and characterization of wild galactose fermenting Streptococcus thermophilus strains. Int. J. Adv. Res. 1 83–96.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

