Insights into the Binding Recognition and Susceptibility of Tofacitinib toward Janus Kinases
- PMID: 31956784
- PMCID: PMC6964278
- DOI: 10.1021/acsomega.9b02800
Insights into the Binding Recognition and Susceptibility of Tofacitinib toward Janus Kinases
Abstract
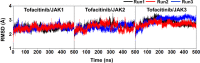
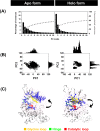
Janus kinases (JAKs) are enzymes involved in signaling pathways that affect hematopoiesis and immune cell functions. JAK1, JAK2, and JAK3 play different roles in numerous diseases of the immune system and have also been considered as potential targets for cancer therapy. In the present study, the susceptibility of the oral JAK inhibitor tofacitinib against these three JAKs was elucidated using the 500-ns molecular dynamics (MD) simulations and free energy calculations based on MM-PB(GB)SA, QM/MM-GBSA (PM3 and SCC-DFTB), and SIE methods. The obtained results revealed that tofacitinib could interact with all JAKs at the ATP-binding site via electrostatic attraction, hydrogen bond formation, and in particular van der Waals interaction. The conserved glutamate and leucine residues (E957 and L959 of JAK1, E930 and L932 of JAK2, and E903 and L905 of JAK3) located in the hinge region stabilized tofacitinib binding through strongly formed hydrogen bonds. Complexation with the incoming tofacitinib led to a closed conformation of the ATP-binding site and a decreased protein fluctuation at the glycine loop of the JAK protein. The binding affinities of tofacitinib/JAKs were ranked in the order of JAK3 > JAK2 ∼ JAK1, which are in line with the reported experimental data.
Copyright © 2020 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
