Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium
- PMID: 31956913
- PMCID: PMC7102962
- DOI: 10.1093/nar/gkaa012
Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium
Abstract
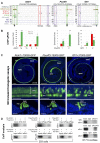
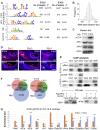
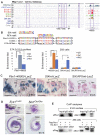
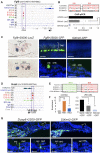
The transcription factor Six1 is essential for induction of sensory cell fate and formation of auditory sensory epithelium, but how it activates gene expression programs to generate distinct cell-types remains unknown. Here, we perform genome-wide characterization of Six1 binding at different stages of auditory sensory epithelium development and find that Six1-binding to cis-regulatory elements changes dramatically at cell-state transitions. Intriguingly, Six1 pre-occupies enhancers of cell-type-specific regulators and effectors before their expression. We demonstrate in-vivo cell-type-specific activity of Six1-bound novel enhancers of Pbx1, Fgf8, Dusp6, Vangl2, the hair-cell master regulator Atoh1 and a cascade of Atoh1's downstream factors, including Pou4f3 and Gfi1. A subset of Six1-bound sites carry consensus-sequences for its downstream factors, including Atoh1, Gfi1, Pou4f3, Gata3 and Pbx1, all of which physically interact with Six1. Motif analysis identifies RFX/X-box as one of the most significantly enriched motifs in Six1-bound sites, and we demonstrate that Six1-RFX proteins cooperatively regulate gene expression through binding to SIX:RFX-motifs. Six1 targets a wide range of hair-bundle regulators and late Six1 deletion disrupts hair-bundle polarity. This study provides a mechanistic understanding of how Six1 cooperates with distinct cofactors in feedforward loops to control lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Oliver G., Wehr R., Jenkins N. A., Copeland N. G., Cheyette B. N., Hartenstein V., Zipursky S. L., Gruss P.. Homeobox genes and connective tissue patterning. Development. 1995; 121:693–705. - PubMed
-
- Kingsbury T.J., Kim M., Civin C.I.. Regulation of cancer stem cell properties by SIX1, a member of the PAX-SIX-EYA-DACH network. Adv. Cancer Res. 2019; 141:1–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

