Genetic diagnoses in epilepsy: The impact of dynamic exome analysis in a pediatric cohort
- PMID: 31957018
- PMCID: PMC7404709
- DOI: 10.1111/epi.16427
Genetic diagnoses in epilepsy: The impact of dynamic exome analysis in a pediatric cohort
Abstract
Objective: We evaluated the yield of systematic analysis and/or reanalysis of whole exome sequencing (WES) data from a cohort of well-phenotyped pediatric patients with epilepsy and suspected but previously undetermined genetic etiology.
Methods: We identified and phenotyped 125 participants with pediatric epilepsy. Etiology was unexplained at the time of enrollment despite clinical testing, which included chromosomal microarray (57 patients), epilepsy gene panel (n = 48), both (n = 28), or WES (n = 8). Clinical epilepsy diagnoses included developmental and epileptic encephalopathy (DEE), febrile infection-related epilepsy syndrome, Rasmussen encephalitis, and other focal and generalized epilepsies. We analyzed WES data and compared the yield in participants with and without prior clinical genetic testing.
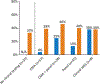
Results: Overall, we identified pathogenic or likely pathogenic variants in 40% (50/125) of our study participants. Nine patients with DEE had genetic variants in recently published genes that had not been recognized as epilepsy-related at the time of clinical testing (FGF12, GABBR1, GABBR2, ITPA, KAT6A, PTPN23, RHOBTB2, SATB2), and eight patients had genetic variants in candidate epilepsy genes (CAMTA1, FAT3, GABRA6, HUWE1, PTCHD1). Ninety participants had concomitant or subsequent clinical genetic testing, which was ultimately explanatory for 26% (23/90). Of the 67 participants whose molecular diagnoses were "unsolved" through clinical genetic testing, we identified pathogenic or likely pathogenic variants in 17 (25%).
Significance: Our data argue for early consideration of WES with iterative reanalysis for patients with epilepsy, particularly those with DEE or epilepsy with intellectual disability. Rigorous analysis of WES data of well-phenotyped patients with epilepsy leads to a broader understanding of gene-specific phenotypic spectra as well as candidate disease gene identification. We illustrate the dynamic nature of genetic diagnosis over time, with analysis and in some cases reanalysis of exome data leading to the identification of disease-associated variants among participants with previously nondiagnostic results from a variety of clinical testing strategies.
Keywords: epilepsy; genetics; phenotype; reanalysis; whole exome sequencing.
Wiley Periodicals, Inc. © 2020 International League Against Epilepsy.
Conflict of interest statement
Conflicts of Interest
Nothing to report. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Figures
References
-
- Kevelam SH, Bierau J, Salvarinova R, et al. Recessive ITPA mutations cause an early infantile encephalopathy. Ann Neurol. 2015; 78(4):649–58. - PubMed
-
- Nambot S, Thevenon J, Kuentz P, et al. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and / or intellectual disability : substantial interest of prospective annual reanalysis. Nat Publ Gr [Internet]. 2017; 20(6):645–54. Available from: 10.1038/gim.2017.162 - DOI - PubMed
-
- Hildebrand MS, Dahl H-HM, Damiano JA, et al. Recent advances in the molecular genetics of epilepsy. J Med Genet [Internet]. 2013; 50(5):271–9. Available from: http://jmg.bmj.com/lookup/doi/10.1136/jmedgenet-2012-101448 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

