Combined analysis of whole-exon sequencing and lncRNA sequencing in type 2 diabetes mellitus patients with obesity
- PMID: 31957265
- PMCID: PMC7028848
- DOI: 10.1111/jcmm.14932
Combined analysis of whole-exon sequencing and lncRNA sequencing in type 2 diabetes mellitus patients with obesity
Abstract
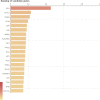
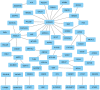
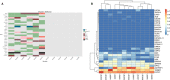
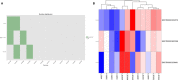
This study sought to find more exon mutation sites and lncRNA candidates associated with type 2 diabetes mellitus (T2DM) patients with obesity (O-T2DM). We used O-T2DM patients and healthy individuals to detect mutations in their peripheral blood by whole-exon sequencing. And changes in lncRNA expression caused by mutation sites were studied at the RNA level. Then, we performed GO analysis and KEGG pathway analysis. We found a total of 277 377 mutation sites between O-T2DM and healthy individuals. Then, we performed a DNA-RNA joint analysis. Based on the screening of harmful sites, 30 mutant genes shared in O-T2DM patients were screened. At the RNA level, mutations of 106 differentially expressed genes were displayed. Finally, a consensus mutation site and differential expression consensus gene screening were performed. In the current study, the results revealed significant differences in exon sites in peripheral blood between O-T2DM and healthy individuals, which may play an important role in the pathogenesis of O-T2DM by affecting the expression of the corresponding lncRNA. This study provides clues to the molecular mechanisms of metabolic disorders in O-T2DM patients at the DNA and RNA levels, as well as biomarkers of the risk of these disorders.
Keywords: diabetes; long non-coding RNA; mutation sites; obesity.
© 2020 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures



Similar articles
-
The Construction and Analysis of the Aberrant lncRNA-miRNA-mRNA Network in Adipose Tissue from Type 2 Diabetes Individuals with Obesity.J Diabetes Res. 2020 Apr 8;2020:3980742. doi: 10.1155/2020/3980742. eCollection 2020. J Diabetes Res. 2020. PMID: 32337289 Free PMC article.
-
Long non-coding RNA LY86-AS1 and HCG27_201 expression in type 2 diabetes mellitus.Mol Biol Rep. 2018 Dec;45(6):2601-2608. doi: 10.1007/s11033-018-4429-8. Epub 2018 Oct 16. Mol Biol Rep. 2018. PMID: 30328000
-
Microarray Profiling and Coexpression Network Analysis of Long Noncoding RNAs in Adipose Tissue of Obesity-T2DM Mouse.Obesity (Silver Spring). 2019 Oct;27(10):1644-1651. doi: 10.1002/oby.22590. Epub 2019 Aug 29. Obesity (Silver Spring). 2019. PMID: 31464075
-
Identification of Potential Therapeutic Targets and Pathways of Liraglutide Against Type 2 Diabetes Mellitus (T2DM) Based on Long Non-Coding RNA (lncRNA) Sequencing.Med Sci Monit. 2020 Apr 2;26:e922210. doi: 10.12659/MSM.922210. Med Sci Monit. 2020. PMID: 32238798 Free PMC article.
-
Combined analysis of whole-exome sequencing and RNA sequencing in type 2 diabetes mellitus patients with thirst and fatigue.Diabetol Metab Syndr. 2022 Aug 8;14(1):111. doi: 10.1186/s13098-022-00884-z. Diabetol Metab Syndr. 2022. PMID: 35941691 Free PMC article.
Cited by
-
Identification of a Multi-Messenger RNA Signature as Type 2 Diabetes Mellitus Candidate Genes Involved in Crosstalk between Inflammation and Insulin Resistance.Biomolecules. 2022 Sep 2;12(9):1230. doi: 10.3390/biom12091230. Biomolecules. 2022. PMID: 36139069 Free PMC article.
-
Long non-coding RNA screening and identification of potential biomarkers for type 2 diabetes.J Clin Lab Anal. 2022 Apr;36(4):e24280. doi: 10.1002/jcla.24280. Epub 2022 Mar 7. J Clin Lab Anal. 2022. PMID: 35257412 Free PMC article.
-
Noncoding RNA profiling in omentum adipose tissue from obese patients and the identification of novel metabolic biomarkers.Front Genet. 2025 Feb 6;16:1533637. doi: 10.3389/fgene.2025.1533637. eCollection 2025. Front Genet. 2025. PMID: 39981261 Free PMC article.
-
Identification of novel genes associated with atherosclerosis in Bama miniature pig.Animal Model Exp Med. 2024 Jun;7(3):377-387. doi: 10.1002/ame2.12412. Epub 2024 May 8. Animal Model Exp Med. 2024. PMID: 38720469 Free PMC article.
-
Machine learning-based stratification of prediabetes and type 2 diabetes progression.Diabetol Metab Syndr. 2025 Jun 18;17(1):227. doi: 10.1186/s13098-025-01786-6. Diabetol Metab Syndr. 2025. PMID: 40533788 Free PMC article.
References
-
- Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13(8):565‐575. - PubMed
-
- Xiaowei Z, Huifang L, Diabetology DO. Application of whole genome exon sequencing in type 2 diabetes mellitus. Chinese J Clin (Electronic Edition). 2018;12(6):46‐49.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

