Setting up for gastrulation: D. melanogaster
- PMID: 31959292
- PMCID: PMC7296044
- DOI: 10.1016/bs.ctdb.2019.11.004
Setting up for gastrulation: D. melanogaster
Abstract
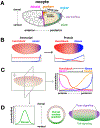
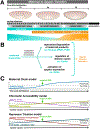
Drosophila melanogaster embryos develop initially as a syncytium of totipotent nuclei and subsequently, once cellularized, undergo morphogenetic movements associated with gastrulation to generate the three somatic germ layers of the embryo: mesoderm, ectoderm, and endoderm. In this chapter, we focus on the first phase of gastrulation in Drosophila involving patterning of early embryos when cells differentiate their gene expression programs. This patterning process requires coordination of multiple developmental processes including genome reprogramming at the maternal-to-zygotic transition, combinatorial action of transcription factors to support distinct gene expression, and dynamic feedback between this genetic patterning by transcription factors and changes in cell morphology. We discuss the gene regulatory programs acting during patterning to specify the three germ layers, which involve the regulation of spatiotemporal gene expression coupled to physical tissue morphogenesis.
Keywords: Anterior-posterior patterning; Dorsal-ventral patterning; Drosophila melanogaster; Ectoderm; Embryonic development; Endoderm; Gastrulation; Germ-band elongation; Maternal-to-zygotic transition; Mesoderm; Morphogen gradients; Syncytium.
© 2020 Elsevier Inc. All rights reserved.
Figures


References
-
- Alberga A, Boulay JL, Kempe E, Dennefeld C, & Haenlin M (1991). The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development, 111(4), 983–992. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

