A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin
- PMID: 31959557
- PMCID: PMC7571800
- DOI: 10.1016/j.molcel.2019.12.019
A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin
Abstract
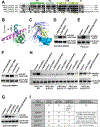
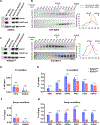
Diverse accessory subunits are involved in the recruitment of polycomb repressive complex 2 (PRC2) to CpG island (CGI) chromatin. Here we report the crystal structure of a SUZ12-RBBP4 complex bound to fragments of the accessory subunits PHF19 and JARID2. Unexpectedly, this complex adopts a dimeric structural architecture, accounting for PRC2 self-association that has long been implicated. The intrinsic PRC2 dimer is formed via domain swapping involving RBBP4 and the unique C2 domain of SUZ12. MTF2 and PHF19 associate with PRC2 at around the dimer interface and stabilize the dimer. Conversely, AEBP2 binding results in a drastic movement of the C2 domain, disrupting the intrinsic PRC2 dimer. PRC2 dimerization enhances CGI DNA binding by PCLs in pairs in vitro, reminiscent of the widespread phenomenon of transcription factor dimerization in active transcription. Loss of PRC2 dimerization impairs histone H3K27 trimethylation (H3K27me3) on chromatin at developmental gene loci in mouse embryonic stem cells.
Keywords: CpG island (CGI); chromatin; crystal structure; epigenetics; gene silencing; histone methylation; polycomb repressive complex 2 (PRC2); polycomb-like (PCL); protein dimerization; structural biology.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





Similar articles
-
Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.Mol Cell. 2018 Mar 1;69(5):840-852.e5. doi: 10.1016/j.molcel.2018.01.039. Mol Cell. 2018. PMID: 29499137 Free PMC article.
-
Polycomb-like proteins link the PRC2 complex to CpG islands.Nature. 2017 Sep 14;549(7671):287-291. doi: 10.1038/nature23881. Epub 2017 Sep 6. Nature. 2017. PMID: 28869966 Free PMC article.
-
PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation.Mol Cell. 2019 Nov 7;76(3):437-452.e6. doi: 10.1016/j.molcel.2019.08.012. Epub 2019 Sep 11. Mol Cell. 2019. PMID: 31521505
-
A Structural Perspective on Gene Repression by Polycomb Repressive Complex 2.Subcell Biochem. 2021;96:519-562. doi: 10.1007/978-3-030-58971-4_17. Subcell Biochem. 2021. PMID: 33252743 Review.
-
Tissue-Specific Tumour Suppressor and Oncogenic Activities of the Polycomb-like Protein MTF2.Genes (Basel). 2023 Sep 27;14(10):1879. doi: 10.3390/genes14101879. Genes (Basel). 2023. PMID: 37895228 Free PMC article. Review.
Cited by
-
Critical Roles of Polycomb Repressive Complexes in Transcription and Cancer.Int J Mol Sci. 2022 Aug 24;23(17):9574. doi: 10.3390/ijms23179574. Int J Mol Sci. 2022. PMID: 36076977 Free PMC article. Review.
-
The molecular principles of gene regulation by Polycomb repressive complexes.Nat Rev Mol Cell Biol. 2021 Dec;22(12):815-833. doi: 10.1038/s41580-021-00398-y. Epub 2021 Aug 16. Nat Rev Mol Cell Biol. 2021. PMID: 34400841 Free PMC article. Review.
-
Cooperative DNA looping by PRC2 complexes.Nucleic Acids Res. 2021 Jun 21;49(11):6238-6248. doi: 10.1093/nar/gkab441. Nucleic Acids Res. 2021. PMID: 34057467 Free PMC article.
-
Unveiling the molecular structure and role of RBBP4/7: implications for epigenetic regulation and cancer research.Front Mol Biosci. 2023 Nov 13;10:1276612. doi: 10.3389/fmolb.2023.1276612. eCollection 2023. Front Mol Biosci. 2023. PMID: 38028543 Free PMC article. Review.
-
Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.Elife. 2020 Nov 19;9:e61964. doi: 10.7554/eLife.61964. Elife. 2020. PMID: 33211010 Free PMC article.
References
-
- Amoutzias GD, Robertson DL, Van de Peer Y, and Oliver SG (2008). Choose your partners: dimerization in eukaryotic transcription factors. Trends in biochemical sciences 33, 220–229. - PubMed
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. - PubMed
-
- Cao R, and Zhang Y (2004). SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Molecular cell 15, 57–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

