Extracellular NAD+ enhances PARP-dependent DNA repair capacity independently of CD73 activity
- PMID: 31959836
- PMCID: PMC6971268
- DOI: 10.1038/s41598-020-57506-9
Extracellular NAD+ enhances PARP-dependent DNA repair capacity independently of CD73 activity
Abstract
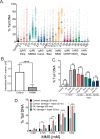
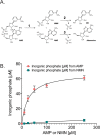
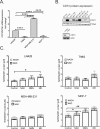
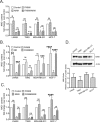
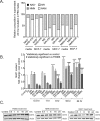
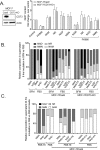
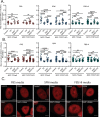
Changes in nicotinamide adenine dinucleotide (NAD+) levels that compromise mitochondrial function trigger release of DNA damaging reactive oxygen species. NAD+ levels also affect DNA repair capacity as NAD+ is a substrate for PARP-enzymes (mono/poly-ADP-ribosylation) and sirtuins (deacetylation). The ecto-5'-nucleotidase CD73, an ectoenzyme highly expressed in cancer, is suggested to regulate intracellular NAD+ levels by processing NAD+ and its bio-precursor, nicotinamide mononucleotide (NMN), from tumor microenvironments, thereby enhancing tumor DNA repair capacity and chemotherapy resistance. We therefore investigated whether expression of CD73 impacts intracellular NAD+ content and NAD+-dependent DNA repair capacity. Reduced intracellular NAD+ levels suppressed recruitment of the DNA repair protein XRCC1 to sites of genomic DNA damage and impacted the amount of accumulated DNA damage. Further, decreased NAD+ reduced the capacity to repair DNA damage induced by DNA alkylating agents. Overall, reversal of these outcomes through NAD+ or NMN supplementation was independent of CD73. In opposition to its proposed role in extracellular NAD+ bioprocessing, we found that recombinant human CD73 only poorly processes NMN but not NAD+. A positive correlation between CD73 expression and intracellular NAD+ content could not be made as CD73 knockout human cells were efficient in generating intracellular NAD+ when supplemented with NAD+ or NMN.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

