Eighty-eight variants highlight the role of T cell regulation and airway remodeling in asthma pathogenesis
- PMID: 31959851
- PMCID: PMC6971247
- DOI: 10.1038/s41467-019-14144-8
Eighty-eight variants highlight the role of T cell regulation and airway remodeling in asthma pathogenesis
Abstract
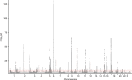
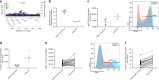
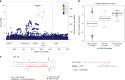
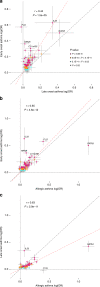
Asthma is one of the most common chronic diseases affecting both children and adults. We report a genome-wide association meta-analysis of 69,189 cases and 702,199 controls from Iceland and UK biobank. We find 88 asthma risk variants at 56 loci, 19 previously unreported, and evaluate their effect on other asthma and allergic phenotypes. Of special interest are two low frequency variants associated with protection against asthma; a missense variant in TNFRSF8 and 3' UTR variant in TGFBR1. Functional studies show that the TNFRSF8 variant reduces TNFRSF8 expression both on cell surface and in soluble form, acting as loss of function. eQTL analysis suggests that the TGFBR1 variant acts through gain of function and together with an intronic variant in a downstream gene, SMAD3, points to defective TGFβR1 signaling as one of the biological perturbations increasing asthma risk. Our results increase the number of asthma variants and implicate genes with known role in T cell regulation, inflammation and airway remodeling in asthma pathogenesis.
Conflict of interest statement
Authors affiliated with deCODE genetics/Amgen Inc. declare competing interests as employees. The remaining authors declare no competing interests.
Figures