A novel pathway regulates social hierarchy via lncRNA AtLAS and postsynaptic synapsin IIb
- PMID: 31959917
- PMCID: PMC7015055
- DOI: 10.1038/s41422-020-0273-1
A novel pathway regulates social hierarchy via lncRNA AtLAS and postsynaptic synapsin IIb
Erratum in
-
Author Correction: A novel pathway regulates social hierarchy via lncRNA AtLAS and postsynaptic synapsin IIb.Cell Res. 2021 May;31(5):601. doi: 10.1038/s41422-021-00492-y. Cell Res. 2021. PMID: 33707743 Free PMC article. No abstract available.
Abstract
Dominance hierarchy is a fundamental phenomenon in grouped animals and human beings, however, the underlying regulatory mechanisms remain elusive. Here, we report that an antisense long non-coding RNA (lncRNA) of synapsin II, named as AtLAS, plays a crucial role in the regulation of social hierarchy. AtLAS is decreased in the prefrontal cortical excitatory pyramidal neurons of dominant mice; consistently, silencing or overexpression of AtLAS increases or decreases the social rank, respectively. Mechanistically, we show that AtLAS regulates alternative polyadenylation of synapsin II gene and increases synapsin 2b (syn2b) expression. Syn2b reduces AMPA receptor (AMPAR)-mediated excitatory synaptic transmission through a direct binding with AMPAR at the postsynaptic site via its unique C-terminal sequence. Moreover, a peptide disrupting the binding of syn2b with AMPARs enhances the synaptic strength and social ranks. These findings reveal a novel role for lncRNA AtLAS and its target syn2b in the regulation of social behaviors by controlling postsynaptic AMPAR trafficking.
Conflict of interest statement
The peptide used to block the binding of syn2b with AMPAR has been submitted to the Patent Office of the Peopleʼs Republic of China by L.Q.Z., D.L., W.X., M.M., X.H. (Application No. 201811638624.0). No other conflicts of interests are declared for all authors.
Figures
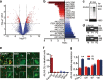

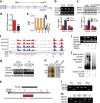

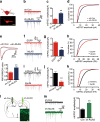
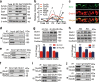

Comment in
-
Expanding the AtLAS of non-coding RNA functions in the brain.Cell Res. 2020 Apr;30(4):283-284. doi: 10.1038/s41422-020-0289-6. Cell Res. 2020. PMID: 32111970 Free PMC article. No abstract available.
Similar articles
-
Synapsin Isoforms Regulating GABA Release from Hippocampal Interneurons.J Neurosci. 2016 Jun 22;36(25):6742-57. doi: 10.1523/JNEUROSCI.0011-16.2016. J Neurosci. 2016. PMID: 27335405 Free PMC article.
-
AMPA receptor synaptic targeting regulated by stargazin interactions with the Golgi-resident PDZ protein nPIST.J Neurosci. 2004 Aug 25;24(34):7491-502. doi: 10.1523/JNEUROSCI.1255-04.2004. J Neurosci. 2004. PMID: 15329396 Free PMC article.
-
SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons.Proc Natl Acad Sci U S A. 2006 Mar 21;103(12):4344-51. doi: 10.1073/pnas.0600084103. Epub 2006 Mar 14. Proc Natl Acad Sci U S A. 2006. PMID: 16537406 Free PMC article.
-
Regulation of neuronal PKA signaling through AKAP targeting dynamics.Eur J Cell Biol. 2006 Jul;85(7):627-33. doi: 10.1016/j.ejcb.2006.01.010. Epub 2006 Feb 28. Eur J Cell Biol. 2006. PMID: 16504338 Review.
-
Glutamatergic neurotransmission in the synapsin I and II double knock-out mouse.Semin Cell Dev Biol. 2011 Jun;22(4):400-7. doi: 10.1016/j.semcdb.2011.07.004. Epub 2011 Jul 31. Semin Cell Dev Biol. 2011. PMID: 21827868 Review.
Cited by
-
Correlated evolution of social organization and lifespan in mammals.Nat Commun. 2023 Jan 31;14(1):372. doi: 10.1038/s41467-023-35869-7. Nat Commun. 2023. PMID: 36720880 Free PMC article.
-
Sociability genetically separable from social hierarchy in amniotes.iScience. 2025 Jun 18;28(7):112930. doi: 10.1016/j.isci.2025.112930. eCollection 2025 Jul 18. iScience. 2025. PMID: 40678513 Free PMC article.
-
Senktide blocks aberrant RTN3 interactome to retard memory decline and tau pathology in social isolated Alzheimer's disease mice.Protein Cell. 2024 Apr 1;15(4):261-284. doi: 10.1093/procel/pwad056. Protein Cell. 2024. PMID: 38011644 Free PMC article.
-
Natural antisense transcripts as drug targets.Front Mol Biosci. 2022 Sep 27;9:978375. doi: 10.3389/fmolb.2022.978375. eCollection 2022. Front Mol Biosci. 2022. PMID: 36250017 Free PMC article. Review.
-
Current and Future Perspectives of Noncoding RNAs in Brain Function and Neuropsychiatric Disease.Biol Psychiatry. 2022 Jan 15;91(2):183-193. doi: 10.1016/j.biopsych.2021.08.013. Epub 2021 Aug 24. Biol Psychiatry. 2022. PMID: 34742545 Free PMC article. Review.
References
-
- Sapolsky RM. The influence of social hierarchy on primate health. Science. 2005;308:648–652. - PubMed
-
- Nagy M, Akos Z, Biro D, Vicsek T. Hierarchical group dynamics in pigeon flocks. Nature. 2010;464:890–893. - PubMed
-
- Singer T. The past, present and future of social neuroscience: a European perspective. NeuroImage. 2012;61:437–449. - PubMed
-
- Wang F, et al. Bidirectional control of social hierarchy by synaptic efficacy in medial prefrontal cortex. Science. 2011;334:693–697. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

