Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain
- PMID: 31959936
- PMCID: PMC7595134
- DOI: 10.1038/s41593-019-0566-1
Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain
Erratum in
-
Author Correction: Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain.Nat Neurosci. 2020 Feb;23(2):294. doi: 10.1038/s41593-020-0595-9. Nat Neurosci. 2020. PMID: 32005940
-
Author Correction: Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain.Nat Neurosci. 2020 Oct;23(10):1308. doi: 10.1038/s41593-020-0682-y. Nat Neurosci. 2020. PMID: 32719564
Abstract
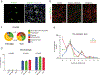
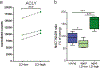
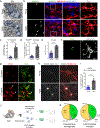
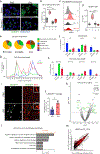
Microglia become progressively activated and seemingly dysfunctional with age, and genetic studies have linked these cells to the pathogenesis of a growing number of neurodegenerative diseases. Here we report a striking buildup of lipid droplets in microglia with aging in mouse and human brains. These cells, which we call 'lipid-droplet-accumulating microglia' (LDAM), are defective in phagocytosis, produce high levels of reactive oxygen species and secrete proinflammatory cytokines. RNA-sequencing analysis of LDAM revealed a transcriptional profile driven by innate inflammation that is distinct from previously reported microglial states. An unbiased CRISPR-Cas9 screen identified genetic modifiers of lipid droplet formation; surprisingly, variants of several of these genes, including progranulin (GRN), are causes of autosomal-dominant forms of human neurodegenerative diseases. We therefore propose that LDAM contribute to age-related and genetic forms of neurodegeneration.
Figures









Comment in
-
New Microglia on the Block.Cell Metab. 2020 Apr 7;31(4):664-666. doi: 10.1016/j.cmet.2020.03.015. Cell Metab. 2020. PMID: 32268112
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

