Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1
- PMID: 31959982
- PMCID: PMC6983325
- DOI: 10.1038/s41590-019-0578-8
Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1
Erratum in
-
Author Correction: Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1.Nat Immunol. 2020 Jun;21(6):695. doi: 10.1038/s41590-020-0640-6. Nat Immunol. 2020. PMID: 32123375
Abstract
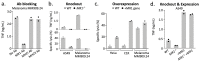
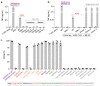
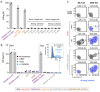
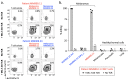
Human leukocyte antigen (HLA)-independent, T cell-mediated targeting of cancer cells would allow immune destruction of malignancies in all individuals. Here, we use genome-wide CRISPR-Cas9 screening to establish that a T cell receptor (TCR) recognized and killed most human cancer types via the monomorphic MHC class I-related protein, MR1, while remaining inert to noncancerous cells. Unlike mucosal-associated invariant T cells, recognition of target cells by the TCR was independent of bacterial loading. Furthermore, concentration-dependent addition of vitamin B-related metabolite ligands of MR1 reduced TCR recognition of cancer cells, suggesting that recognition occurred via sensing of the cancer metabolome. An MR1-restricted T cell clone mediated in vivo regression of leukemia and conferred enhanced survival of NSG mice. TCR transfer to T cells of patients enabled killing of autologous and nonautologous melanoma. These findings offer opportunities for HLA-independent, pan-cancer, pan-population immunotherapies.
Conflict of interest statement
Cardiff University has filed patents based on these findings.
Figures




Comment in
-
'Bohemian Rhapsody' of MR1T cells.Nat Immunol. 2020 Feb;21(2):108-110. doi: 10.1038/s41590-019-0588-6. Nat Immunol. 2020. PMID: 31959981 No abstract available.
-
MR1-restricted pan-cancer T cells.Nat Rev Immunol. 2020 Mar;20(3):141. doi: 10.1038/s41577-020-0284-7. Nat Rev Immunol. 2020. PMID: 32024985 No abstract available.
References
-
- Vavassori S, et al. Butyrophilin 3A1 binds phosphorylated antigens and stimulates human γδ T cells. Nat Immunol. 2013;14:908–916. - PubMed
-
- Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. - PubMed
-
- Corbett AJ, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

