A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance
- PMID: 31959991
- PMCID: PMC7018620
- DOI: 10.1038/s41591-019-0730-x
A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance
Erratum in
-
Author Correction: A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance.Nat Med. 2020 Feb;26(2):300. doi: 10.1038/s41591-020-0773-z. Nat Med. 2020. PMID: 31996837 Free PMC article.
Abstract
Mucociliary clearance, the physiological process by which mammalian conducting airways expel pathogens and unwanted surface materials from the respiratory tract, depends on the coordinated function of multiple specialized cell types, including basal stem cells, mucus-secreting goblet cells, motile ciliated cells, cystic fibrosis transmembrane conductance regulator (CFTR)-rich ionocytes, and immune cells1,2. Bronchiectasis, a syndrome of pathological airway dilation associated with impaired mucociliary clearance, may occur sporadically or as a consequence of Mendelian inheritance, for example in cystic fibrosis, primary ciliary dyskinesia (PCD), and select immunodeficiencies3. Previous studies have identified mutations that affect ciliary structure and nucleation in PCD4, but the regulation of mucociliary transport remains incompletely understood, and therapeutic targets for its modulation are lacking. Here we identify a bronchiectasis syndrome caused by mutations that inactivate NIMA-related kinase 10 (NEK10), a protein kinase with previously unknown in vivo functions in mammals. Genetically modified primary human airway cultures establish NEK10 as a ciliated-cell-specific kinase whose activity regulates the motile ciliary proteome to promote ciliary length and mucociliary transport but which is dispensable for normal ciliary number, radial structure, and beat frequency. Together, these data identify a novel and likely targetable signaling axis that controls motile ciliary function in humans and has potential implications for other respiratory disorders that are characterized by impaired mucociliary clearance.
Conflict of interest statement
Competing Interests Statement
The authors declare the following competing interests:
Provisional patent application in process:
Figures
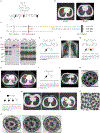

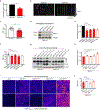





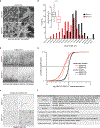
References
Main Text References
-
- Gould CM, Freeman AF & Olivier KN Genetic Causes of Bronchiectasis. Clinics in Chest Medicine 33, 249–263 (2012). - PubMed
-
- Zariwala MA, Knowles MR & Omran H Genetic defects in ciliary structure and function. Annu. Rev. Physiol 69, 423–450 (2007). - PubMed
-
- Online Mendelian Inheritance in Man, OMIM. omim.org Available at: https://www.omim.org/. (Accessed: 11 November 2019)
Methods-only References
-
- Hoffmann K & Lindner TH easyLINKAGE-Plus—automated linkage analyses using large-scale SNP data. Bioinformatics 21, 3565–3567 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U2C TR002818/TR/NCATS NIH HHS/United States
- U54 HL096458/HL/NHLBI NIH HHS/United States
- R01 HL117836/HL/NHLBI NIH HHS/United States
- R01 HL071798/HL/NHLBI NIH HHS/United States
- UL1 TR000083/TR/NCATS NIH HHS/United States
- S10 OD012027/OD/NIH HHS/United States
- K08 HL133603/HL/NHLBI NIH HHS/United States
- R01 AI047389/AI/NIAID NIH HHS/United States
- R01 CA103866/CA/NCI NIH HHS/United States
- T32 CA009216/CA/NCI NIH HHS/United States
- R37 AI047389/AI/NIAID NIH HHS/United States
- T32 HL116275/HL/NHLBI NIH HHS/United States
- R01 CA129105/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

