Building a pipeline to solicit expert knowledge from the community to aid gene summary curation
- PMID: 31960022
- PMCID: PMC6971343
- DOI: 10.1093/database/baz152
Building a pipeline to solicit expert knowledge from the community to aid gene summary curation
Abstract
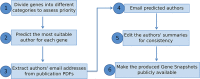
Brief summaries describing the function of each gene's product(s) are of great value to the research community, especially when interpreting genome-wide studies that reveal changes to hundreds of genes. However, manually writing such summaries, even for a single species, is a daunting task; for example, the Drosophila melanogaster genome contains almost 14 000 protein-coding genes. One solution is to use computational methods to generate summaries, but this often fails to capture the key functions or express them eloquently. Here, we describe how we solicited help from the research community to generate manually written summaries of D. melanogaster gene function. Based on the data within the FlyBase database, we developed a computational pipeline to identify researchers who have worked extensively on each gene. We e-mailed these researchers to ask them to draft a brief summary of the main function(s) of the gene's product, which we edited for consistency to produce a 'gene snapshot'. This approach yielded 1800 gene snapshot submissions within a 3-month period. We discuss the general utility of this strategy for other databases that capture data from the research literature. Database URL: https://flybase.org/.
© The Author(s) 2020. Published by Oxford University Press.
Figures



References
-
- Spärck Jones K. (2007) Automatic summarising: The state of the art. Information Processing & Management, 43, 1449.
-
- Jin F., Huang M., Lu Z. and Zhu X. (2009) Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing. Association for Computational Linguistics, Boulder, Colorado, p. 97.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

