Porcine Deltacoronavirus Infection and Transmission in Poultry, United States1
- PMID: 31961296
- PMCID: PMC6986833
- DOI: 10.3201/eid2602.190346
Porcine Deltacoronavirus Infection and Transmission in Poultry, United States1
Abstract
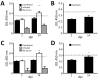
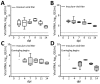
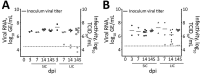
Coronaviruses cause respiratory and gastrointestinal diseases in diverse host species. Deltacoronaviruses (DCoVs) have been identified in various songbird species and in leopard cats in China. In 2009, porcine deltacoronavirus (PDCoV) was detected in fecal samples from pigs in Asia, but its etiologic role was not identified until 2014, when it caused major diarrhea outbreaks in swine in the United States. Studies have shown that PDCoV uses a conserved region of the aminopeptidase N protein to infect cell lines derived from multiple species, including humans, pigs, and chickens. Because PDCoV is a potential zoonotic pathogen, investigations of its prevalence in humans and its contribution to human disease continue. We report experimental PDCoV infection and subsequent transmission among poultry. In PDCoV-inoculated chicks and turkey poults, we observed diarrhea, persistent viral RNA titers from cloacal and tracheal samples, PDCoV-specific serum IgY antibody responses, and antigen-positive cells from intestines.
Keywords: Porcine deltacoronavirus; United States; chickens; coronaviruses; interspecies transmission; poultry; swine disease; turkeys; viruses; zoonoes.
Figures



References
-
- Masters PS, Perlman S. Coronaviridae. In: Knipe DM, Howley PM, eds. Fields virology, 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. p. 825–58.
-
- Schwegmann-Wessels C, Herrler G. Transmissible gastroenteritis virus infection: a vanishing specter. Dtsch Tierarztl Wochenschr. 2006;113:157–9. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

