Oropouche virus cases identified in Ecuador using an optimised qRT-PCR informed by metagenomic sequencing
- PMID: 31961856
- PMCID: PMC6994106
- DOI: 10.1371/journal.pntd.0007897
Oropouche virus cases identified in Ecuador using an optimised qRT-PCR informed by metagenomic sequencing
Abstract
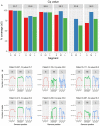
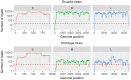
Oropouche virus (OROV) is responsible for outbreaks of Oropouche fever in parts of South America. We recently identified and isolated OROV from a febrile Ecuadorian patient, however, a previously published qRT-PCR assay did not detect OROV in the patient sample. A primer mismatch to the Ecuadorian OROV lineage was identified from metagenomic sequencing data. We report the optimisation of an qRT-PCR assay for the Ecuadorian OROV lineage, which subsequently identified a further five cases in a cohort of 196 febrile patients. We isolated OROV via cell culture and developed an algorithmically-designed primer set for whole-genome amplification of the virus. Metagenomic sequencing of the patient samples provided OROV genome coverage ranging from 68-99%. The additional cases formed a single phylogenetic cluster together with the initial case. OROV should be considered as a differential diagnosis for Ecuadorian patients with febrile illness to avoid mis-diagnosis with other circulating pathogens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Pinheiro FP, Rocha AG, Freitas RB, Ohana BA, Travassos da Rosa AP, Rogério JS, et al. Meningitis associated with Oropouche virus infections. Rev Inst Med Trop Sao Paulo. 1982. July;24(4):246–51. - PubMed

