Virus Metagenomics in Farm Animals: A Systematic Review
- PMID: 31963174
- PMCID: PMC7019290
- DOI: 10.3390/v12010107
Virus Metagenomics in Farm Animals: A Systematic Review
Abstract
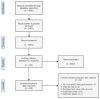
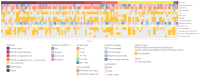
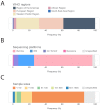
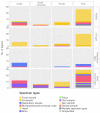
A majority of emerging infectious diseases are of zoonotic origin. Metagenomic Next-Generation Sequencing (mNGS) has been employed to identify uncommon and novel infectious etiologies and characterize virus diversity in human, animal, and environmental samples. Here, we systematically reviewed studies that performed viral mNGS in common livestock (cattle, small ruminants, poultry, and pigs). We identified 2481 records and 120 records were ultimately included after a first and second screening. Pigs were the most frequently studied livestock and the virus diversity found in samples from poultry was the highest. Known animal viruses, zoonotic viruses, and novel viruses were reported in available literature, demonstrating the capacity of mNGS to identify both known and novel viruses. However, the coverage of metagenomic studies was patchy, with few data on the virome of small ruminants and respiratory virome of studied livestock. Essential metadata such as age of livestock and farm types were rarely mentioned in available literature, and only 10.8% of the datasets were publicly available. Developing a deeper understanding of livestock virome is crucial for detection of potential zoonotic and animal pathogens and One Health preparedness. Metagenomic studies can provide this background but only when combined with essential metadata and following the "FAIR" (Findable, Accessible, Interoperable, and Reusable) data principles.
Keywords: NGS; animal reservoir; deep sequencing; emerging infectious diseases; high-throughput sequencing; livestock; one health; viral metagenomics; virome; zoonosis.
Conflict of interest statement
All authors declare no conflicts of interest.
Figures

Similar articles
-
Exploring viral diversity and metagenomics in livestock: insights into disease emergence and spillover risks in cattle.Vet Res Commun. 2024 Aug;48(4):2029-2049. doi: 10.1007/s11259-024-10403-2. Epub 2024 Jun 12. Vet Res Commun. 2024. PMID: 38865041 Review.
-
The Virome of Acute Respiratory Diseases in Individuals at Risk of Zoonotic Infections.Viruses. 2020 Aug 29;12(9):960. doi: 10.3390/v12090960. Viruses. 2020. PMID: 32872469 Free PMC article.
-
Hidden Viral Sequences in Public Sequencing Data and Warning for Future Emerging Diseases.mBio. 2021 Aug 31;12(4):e0163821. doi: 10.1128/mBio.01638-21. Epub 2021 Aug 17. mBio. 2021. PMID: 34399612 Free PMC article.
-
Viral discovery as a tool for pandemic preparedness.Rev Sci Tech. 2017 Aug;36(2):499-512. doi: 10.20506/rst.36.2.2669. Rev Sci Tech. 2017. PMID: 30152468
-
Next generation sequencing technologies: tool to study avian virus diversity.Acta Virol. 2015 Mar;59(1):3-13. doi: 10.4149/av_2015_01_3. Acta Virol. 2015. PMID: 25790045 Review.
Cited by
-
Genome Sequences of Seven Megrivirus Strains from Chickens in The Netherlands.Microbiol Resour Announc. 2020 Nov 19;9(47):e01207-20. doi: 10.1128/MRA.01207-20. Microbiol Resour Announc. 2020. PMID: 33214312 Free PMC article.
-
Conventional diagnosis and metagenomic analysis of a novel co-infection case involving Escherichia coli and immunosuppressive conditions with petechial hepatitis of broilers in South Korea: a case report.BMC Vet Res. 2025 Jul 2;21(1):432. doi: 10.1186/s12917-025-04855-0. BMC Vet Res. 2025. PMID: 40605027 Free PMC article.
-
Potential Use of Microbial Community Genomes in Various Dimensions of Agriculture Productivity and Its Management: A Review.Front Microbiol. 2022 May 17;13:708335. doi: 10.3389/fmicb.2022.708335. eCollection 2022. Front Microbiol. 2022. PMID: 35655999 Free PMC article. Review.
-
Application of shotgun metagenomics sequencing and targeted sequence capture to detect circulating porcine viruses in the Dutch-German border region.Transbound Emerg Dis. 2022 Jul;69(4):2306-2319. doi: 10.1111/tbed.14249. Epub 2021 Aug 28. Transbound Emerg Dis. 2022. PMID: 34347385 Free PMC article.
-
Assessment of Viral Targeted Sequence Capture Using Nanopore Sequencing Directly from Clinical Samples.Viruses. 2020 Nov 27;12(12):1358. doi: 10.3390/v12121358. Viruses. 2020. PMID: 33260903 Free PMC article.
References
-
- Jones B.A., Grace D., Kock R., Alonso S., Rushton J., Said M.Y., McKeever D., Mutua F., Young J., McDermott J., et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. USA. 2013;110:8399–8404. doi: 10.1073/pnas.1208059110. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

