Hybridization Facilitates Adaptive Evolution in Two Major Fungal Pathogens
- PMID: 31963231
- PMCID: PMC7017293
- DOI: 10.3390/genes11010101
Hybridization Facilitates Adaptive Evolution in Two Major Fungal Pathogens
Abstract
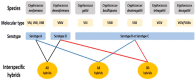
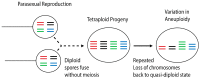
Hybridization is increasingly recognized as an important force impacting adaptation and evolution in many lineages of fungi. During hybridization, divergent genomes and alleles are brought together into the same cell, potentiating adaptation by increasing genomic plasticity. Here, we review hybridization in fungi by focusing on two fungal pathogens of animals. Hybridization is common between the basidiomycete yeast species Cryptococcusneoformans × Cryptococcusdeneoformans, and hybrid genotypes are frequently found in both environmental and clinical settings. The two species show 10-15% nucleotide divergence at the genome level, and their hybrids are highly heterozygous. Though largely sterile and unable to mate, these hybrids can propagate asexually and generate diverse genotypes by nondisjunction, aberrant meiosis, mitotic recombination, and gene conversion. Under stress conditions, the rate of such genetic changes can increase, leading to rapid adaptation. Conversely, in hybrids formed between lineages of the chytridiomycete frog pathogen Batrachochytriumdendrobatidis (Bd), the parental genotypes are considerably less diverged (0.2% divergent). Bd hybrids are formed from crosses between lineages that rarely undergo sex. A common theme in both species is that hybrids show genome plasticity via aneuploidy or loss of heterozygosity and leverage these mechanisms as a rapid way to generate genotypic/phenotypic diversity. Some hybrids show greater fitness and survival in both virulence and virulence-associated phenotypes than parental lineages under certain conditions. These studies showcase how experimentation in model species such as Cryptococcus can be a powerful tool in elucidating the genotypic and phenotypic consequences of hybridization.
Keywords: AD hybrids; Cryptococcus; aneuploidy; frog chytrid; reproductive incompatibilities.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

