Centromeric Non-Coding RNAs: Conservation and Diversity in Function
- PMID: 31963472
- PMCID: PMC7151564
- DOI: 10.3390/ncrna6010004
Centromeric Non-Coding RNAs: Conservation and Diversity in Function
Abstract
Chromosome segregation is strictly regulated for the proper distribution of genetic material to daughter cells. During this process, mitotic chromosomes are pulled to both poles by bundles of microtubules attached to kinetochores that are assembled on the chromosomes. Centromeres are specific regions where kinetochores assemble. Although these regions were previously considered to be silent, some experimental studies have demonstrated that transcription occurs in these regions to generate non-coding RNAs (ncRNAs). These centromeric ncRNAs (cenRNAs) are involved in centromere functions. Here, we describe the currently available information on the functions of cenRNAs in several species.
Keywords: RNP; centromere; chromosome segregation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
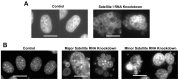
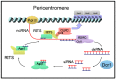

References
Publication types
LinkOut - more resources
Full Text Sources

