Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response
- PMID: 31964396
- PMCID: PMC6971890
- DOI: 10.1186/s12943-019-1122-z
Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response
Abstract
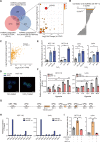
Background: Hypoxic tumors are refractory to DNA damage drugs. However, the underlying mechanism has yet to be elucidated. We aimed to identify lncRNAs that upregulated under hypoxia and their effects on colorectal cancer (CRC).
Methods: CRC cells were treated with 1% O2 to identify lncRNAs that upregulated under hypoxia. We integrated these lncRNAs with RNA-seq of 4 paired CRC tissues and TCGA data to get candidate lncRNAs. Multiple in vitro and in vivo assays were used to explore the role of LUCAT1 in CRC.
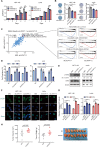
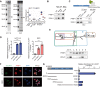
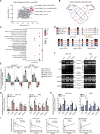
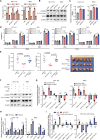
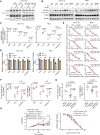
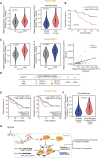
Results: We identified a hypoxia-induced lncRNA LUCAT1 that facilitated the growth of CRC cells and contributed to drug resistance of CRC cells both in vitro and in vivo. Mechanically, LUCAT1 interacts with polypyrimidine tract binding protein 1 (PTBP1) in CRC cells, facilitates the association of a set of DNA damage related genes with PTBP1, thus resulting in altered alternative splicing of these genes. Moreover, ectopic expression of PTBP1 in CRC cells with knockdown of LUCAT1 abrogated the effects induced by LUCAT1 knockdown. Chemotherapeutics drug combined with LUCAT1 knockdown via antisense oligonucleotides (ASO) would get a better outcome in vivo, compared with group treated with chemotherapeutic drug only. Notably, LUCAT1 is upregulated in CRC tissues, compared to adjacent normal tissues; and CRC patients with higher LUCAT1 have a worse prognosis and poorly responded to chemotherapy in the clinic.
Conclusions: Our data suggested CRC cells utilizes LUCAT1 to develop resistance to DNA damage drugs, and disrupting the LUCAT1/PTBP1 axis might be a promising therapeutic strategy for refractory hypoxic tumors.
Keywords: Alternative splicing; Chemoresistance; Hypoxia; LUCAT1; PTBP1; lncRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Leith JT, Padfield G, Faulkner L, Michelson S. Hypoxic fractions in xenografted human colon tumors. Cancer Res. 1991;51:5139–5143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

