Pathobiological Origins and Evolutionary History of Highly Pathogenic Avian Influenza Viruses
- PMID: 31964650
- PMCID: PMC7849344
- DOI: 10.1101/cshperspect.a038679
Pathobiological Origins and Evolutionary History of Highly Pathogenic Avian Influenza Viruses
Abstract
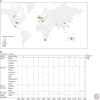
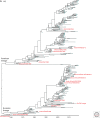
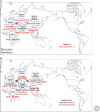
High-pathogenicity avian influenza (HPAI) viruses have arisen from low-pathogenicity avian influenza (LPAI) viruses via changes in the hemagglutinin proteolytic cleavage site, which include mutation of multiple nonbasic to basic amino acids, duplication of basic amino acids, or recombination with insertion of cellular or viral amino acids. Between 1959 and 2019, a total of 42 natural, independent H5 (n = 15) and H7 (n = 27) LPAI to HPAI virus conversion events have occurred in Europe (n = 16), North America (n = 9), Oceania (n = 7), Asia (n = 5), Africa (n = 4), and South America (n = 1). Thirty-eight of these HPAI outbreaks were limited in the number of poultry premises affected and were eradicated. However, poultry outbreaks caused by A/goose/Guangdong/1/1996 (H5Nx), Mexican H7N3, and Chinese H7N9 HPAI lineages have continued. Active surveillance and molecular detection and characterization efforts will provide the best opportunity for early detection and eradication from domestic birds.
Copyright © 2021 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
-
- Abdelwhab EM, Veits J, Tauscher K, Ziller M, Grund C, Hassan MK, Shaheen M, Harder TC, Teifke J, Stech J, et al. 2016. Progressive glycosylation of the haemagglutinin of avian influenza H5N1 modulates virus replication, virulence and chicken-to-chicken transmission without significant impact on antigenic drift. J Gen Virol 97: 3193–3204. 10.1099/jgv.0.000648 - DOI - PubMed
-
- Abolnik C, Londt BZ, Manvell RJ, Shell W, Banks J, Gerdes GH, Akol G, Brown IH. 2009. Characterisation of a highly pathogenic influenza A virus of subtype H5N2 isolated from ostriches in South Africa in 2004. Influenza Other Respir Viruses 3: 63–68. 10.1111/j.1750-2659.2009.00074.x - DOI - PMC - PubMed
-
- Alexander DJ. 2000. The history of avian influenza in poultry. World Poultry Nov: 7–8.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
