Microbiomes Reduce Their Host's Sensitivity to Interspecific Interactions
- PMID: 31964727
- PMCID: PMC6974562
- DOI: 10.1128/mBio.02657-19
Microbiomes Reduce Their Host's Sensitivity to Interspecific Interactions
Abstract
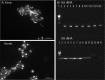
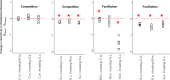
Bacteria associated with eukaryotic hosts can affect host fitness and trophic interactions between eukaryotes, but the extent to which bacteria influence the eukaryotic species interactions within trophic levels that modulate biodiversity and species coexistence is mostly unknown. Here, we used phytoplankton, which are a classic model for evaluating interactions between species, grown with and without associated bacteria to test whether the bacteria alter the strength and type of species interactions within a trophic level. We demonstrate that host-associated bacteria alter host growth rates and carrying capacity. This did not change the type but frequently changed the strength of host interspecific interactions by facilitating host growth in the presence of an established species. These findings indicate that microbiomes can regulate their host species' interspecific interactions. As between-species interaction strength impacts their ability to coexist, our findings show that microbiomes have the potential to modulate eukaryotic species diversity and community composition.IMPORTANCE Description of the Earth's microbiota has recently undergone a phenomenal expansion that has challenged basic assumptions in many areas of biology, including hominid evolution, human gastrointestinal and neurodevelopmental disorders, and plant adaptation to climate change. By using the classic model system of freshwater phytoplankton that has been drawn upon for numerous foundational theories in ecology, we show that microbiomes, by facilitating their host population, can also influence between-species interactions among their eukaryotic hosts. Between-species interactions, including competition for resources, has been a central tenet in the field of ecology because of its implications for the diversity and composition of communities and how this in turn shapes ecosystem functioning.
Keywords: biodiversity; eukaryotic species interactions; microbiome; species coexistence.
Copyright © 2020 Jackrel et al.
Figures

References
-
- Tilman D. 1997. The influence of functional diversity and composition on ecosystem processes. Science 277:1300–1302. doi:10.1126/science.277.5330.1300. - DOI
-
- Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst 31:343–366. doi:10.1146/annurev.ecolsys.31.1.343. - DOI
-
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B. 2017. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171:1015–1028.e13. doi:10.1016/j.cell.2017.09.016. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

