The CYCLIN-DEPENDENT KINASE Module of the Mediator Complex Promotes Flowering and Reproductive Development in Pea
- PMID: 31964799
- PMCID: PMC7054868
- DOI: 10.1104/pp.19.01173
The CYCLIN-DEPENDENT KINASE Module of the Mediator Complex Promotes Flowering and Reproductive Development in Pea
Abstract
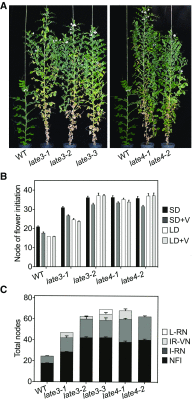
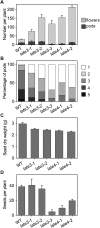
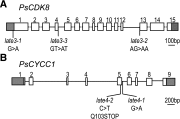
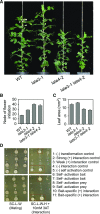
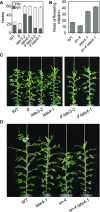
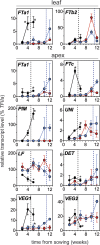
Control of flowering time has been a major focus of comparative genetic analyses in plant development. This study reports on a forward genetic approach to define previously uncharacterized components of flowering control pathways in the long-day legume, pea (Pisum sativum). We isolated two complementation groups of late-flowering mutants in pea that define two uncharacterized loci, LATE BLOOMER3 (LATE3) and LATE4, and describe their diverse effects on vegetative and reproductive development. A map-based comparative approach was employed to identify the underlying genes for both loci, revealing that that LATE3 and LATE4 are orthologs of CYCLIN DEPENDENT KINASE8 (CDK8) and CYCLIN C1 (CYCC1), components of the CDK8 kinase module of the Mediator complex, which is a deeply conserved regulator of transcription in eukaryotes. We confirm the genetic and physical interaction of LATE3 and LATE4 and show that they contribute to the transcriptional regulation of key flowering genes, including the induction of the florigen gene FTa1 and repression of the floral repressor LF Our results establish the conserved importance of the CDK8 module in plants and provide evidence for the function of CYCLIN C1 orthologs in the promotion of flowering and the maintenance of normal reproductive development.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures
References
-
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 - PubMed
-
- Aubert G, Morin J, Jacquin F, Loridon K, Quillet MC, Petit A, Rameau C, Lejeune-Hénaut I, Huguet T, Burstin J (2006) Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor Appl Genet 112: 1024–1041 - PubMed
-
- Berbel A, Ferrándiz C, Hecht V, Dalmais M, Lund OS, Sussmilch FC, Taylor SA, Bendahmane A, Ellis TH, Beltrán JP, et al. (2012) VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat Commun 3: 797. - PubMed
-
- Blümel M, Dally N, Jung C (2015) Flowering time regulation in crops—what did we learn from Arabidopsis? Curr Opin Biotechnol 32: 121–129 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

