Golgi organization is regulated by proteasomal degradation
- PMID: 31964869
- PMCID: PMC6972958
- DOI: 10.1038/s41467-019-14038-9
Golgi organization is regulated by proteasomal degradation
Abstract
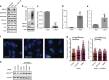
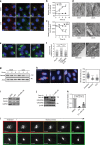
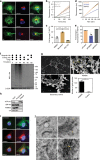
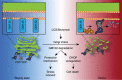
The Golgi is a dynamic organelle whose correct assembly is crucial for cellular homeostasis. Perturbations in Golgi structure are associated with numerous disorders from neurodegeneration to cancer. However, whether and how dispersal of the Golgi apparatus is actively regulated under stress, and the consequences of Golgi dispersal, remain unknown. Here we demonstrate that 26S proteasomes are associated with the cytosolic surface of Golgi membranes to facilitate Golgi Apparatus-Related Degradation (GARD) and degradation of GM130 in response to Golgi stress. The degradation of GM130 is dependent on p97/VCP and 26S proteasomes, and required for Golgi dispersal. Finally, we show that perturbation of Golgi homeostasis induces cell death of multiple myeloma in vitro and in vivo, offering a therapeutic strategy for this malignancy. Taken together, this work reveals a mechanism of Golgi-localized proteasomal degradation, providing a functional link between proteostasis control and Golgi architecture, which may be critical in various secretion-related pathologies.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

