Phase-separation in chromatin organization
- PMID: 31965983
- PMCID: PMC9107952
Phase-separation in chromatin organization
Abstract
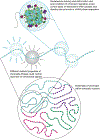
The organization of chromatin into different types of compact versus open states provides a means to fine tune gene regulation. Recent studies have suggested a role for phase-separation in chromatin compaction, raising new possibilities for regulating chromatin compartments. This perspective discusses some specific molecular mechanisms that could leverage such phase-separation processes to control the functions and organization of chromatin.
Figures
Similar articles
-
The Role of Phase Separation in Heterochromatin Formation, Function, and Regulation.Biochemistry. 2018 May 1;57(17):2540-2548. doi: 10.1021/acs.biochem.8b00401. Epub 2018 Apr 23. Biochemistry. 2018. PMID: 29644850 Free PMC article. Review.
-
Multimodal interactions drive chromatin phase separation and compaction.Proc Natl Acad Sci U S A. 2023 Dec 12;120(50):e2308858120. doi: 10.1073/pnas.2308858120. Epub 2023 Dec 4. Proc Natl Acad Sci U S A. 2023. PMID: 38048471 Free PMC article.
-
Chromatin Compaction Leads to a Preference for Peripheral Heterochromatin.Biophys J. 2020 Mar 24;118(6):1479-1488. doi: 10.1016/j.bpj.2020.01.034. Epub 2020 Feb 4. Biophys J. 2020. PMID: 32097622 Free PMC article.
-
HP1 reshapes nucleosome core to promote phase separation of heterochromatin.Nature. 2019 Nov;575(7782):390-394. doi: 10.1038/s41586-019-1669-2. Epub 2019 Oct 16. Nature. 2019. PMID: 31618757 Free PMC article.
-
Chromatin regulation of plant development.Curr Opin Plant Biol. 2003 Feb;6(1):20-8. doi: 10.1016/s1369526602000079. Curr Opin Plant Biol. 2003. PMID: 12495747 Review.
Cited by
-
'RNA modulation of transport properties and stability in phase-separated condensates.Biophys J. 2021 Dec 7;120(23):5169-5186. doi: 10.1016/j.bpj.2021.11.003. Epub 2021 Nov 9. Biophys J. 2021. PMID: 34762868 Free PMC article.
-
OpenABC Enables Flexible, Simplified, and Efficient GPU Accelerated Simulations of Biomolecular Condensates.bioRxiv [Preprint]. 2023 Apr 21:2023.04.19.537533. doi: 10.1101/2023.04.19.537533. bioRxiv. 2023. Update in: PLoS Comput Biol. 2023 Sep 11;19(9):e1011442. doi: 10.1371/journal.pcbi.1011442. PMID: 37131742 Free PMC article. Updated. Preprint.
-
Polymer model integrates imaging and sequencing to reveal how nanoscale heterochromatin domains influence gene expression.Nat Commun. 2025 Apr 23;16(1):3816. doi: 10.1038/s41467-025-59001-z. Nat Commun. 2025. PMID: 40268925 Free PMC article.
-
Chromatin Liquid-Liquid Phase Separation (LLPS) Is Regulated by Ionic Conditions and Fiber Length.Cells. 2022 Oct 6;11(19):3145. doi: 10.3390/cells11193145. Cells. 2022. PMID: 36231107 Free PMC article.
-
Molecular principles of Piwi-mediated cotranscriptional silencing through the dimeric SFiNX complex.Genes Dev. 2021 Mar 1;35(5-6):392-409. doi: 10.1101/gad.347989.120. Epub 2021 Feb 11. Genes Dev. 2021. PMID: 33574069 Free PMC article.
References
-
- Grewal SI and Elgin SC 2002. Heterochromatin new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev 12 178–187 - PubMed
