Site-directed targeting of transcriptional activation-associated proteins to repressed chromatin restores CRISPR activity
- PMID: 31967103
- PMCID: PMC6960031
- DOI: 10.1063/1.5127302
Site-directed targeting of transcriptional activation-associated proteins to repressed chromatin restores CRISPR activity
Abstract
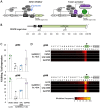
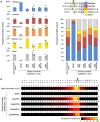
Previously, we used an inducible, transgenic polycomb chromatin system to demonstrate that closed, transcriptionally silenced chromatin reduces Cas9 editing. Here, we investigated strategies to enhance Cas9 editing efficiency by artificially perturbing closed chromatin. We tested UNC1999, a small molecule inhibitor that blocks enhancer of zeste homolog 2, an enzyme that maintains closed polycomb chromatin. We also tested DNA-binding, transiently expressed activation-associated proteins (AAPs) that are known to support an open, transcriptionally active chromatin state. When cells that carried a polycomb-repressed transgene (luciferase) were treated with UNC1999 or the AAP fusion Gal4P65, we observed loss of histone 3 lysine 27 trimethylation (H3K27me3), a silencing-associated chromatin feature, at the transgene. Only Gal4P65 treatment showed full restoration of luciferase expression. Cas9 activity, determined by insertion deletion mutations, was restored in Gal4P65-expressing cells, while no CRISPR enhancement was observed with UNC1999 treatment. CRISPR activity was also restored by other Gal4-AAP fusions that did not activate luciferase expression. Our results demonstrate the use of DNA-binding, activator-associated fusion proteins as an effective method to enhance Cas9 editing within polycomb-repressed chromatin.
© 2020 Author(s).
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

