Physical fitness in community-dwelling older adults is linked to dietary intake, gut microbiota, and metabolomic signatures
- PMID: 31967716
- PMCID: PMC7059135
- DOI: 10.1111/acel.13105
Physical fitness in community-dwelling older adults is linked to dietary intake, gut microbiota, and metabolomic signatures
Abstract
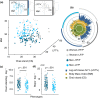
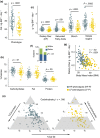
When humans age, changes in body composition arise along with lifestyle-associated disorders influencing fitness and physical decline. Here we provide a comprehensive view of dietary intake, physical activity, gut microbiota (GM), and host metabolome in relation to physical fitness of 207 community-dwelling subjects aged +65 years. Stratification on anthropometric/body composition/physical performance measurements (ABPm) variables identified two phenotypes (high/low-fitness) clearly linked to dietary intake, physical activity, GM, and host metabolome patterns. Strikingly, despite a higher energy intake high-fitness subjects were characterized by leaner bodies and lower fasting proinsulin-C-peptide/blood glucose levels in a mechanism likely driven by higher dietary fiber intake, physical activity and increased abundance of Bifidobacteriales and Clostridiales species in GM and associated metabolites (i.e., enterolactone). These factors explained 50.1% of the individual variation in physical fitness. We propose that targeting dietary strategies for modulation of GM and host metabolome interactions may allow establishing therapeutic approaches to delay and possibly revert comorbidities of aging.
Keywords: aging; energy and dietary fiber intake; gut microbiota; host metabolome; physical fitness; proinsulin-C-peptide.
© 2020 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Aoki, R. , Kamikado, K. , Suda, W. , Takii, H. , Mikami, Y. , Suganuma, N. , … Koga, Y. (2017). A proliferative probiotic Bifidobacterium strain in the gut ameliorates progression of metabolic disorders via microbiota modulation and acetate elevation. Scientific Reports, 7, 43522 10.1038/srep43522 - DOI - PMC - PubMed
-
- Bechshøft, R. L. , Reitelseder, S. , Højfeldt, G. , Castro‐Mejía, J. L. , Khakimov, B. , Ahmad, H. F. B. , … Holm, L. (2016). Counteracting Age‐related Loss of Skeletal Muscle Mass: A clinical and ethnological trial on the role of protein supplementation and training load (CALM Intervention Study): Study protocol for a randomized controlled trial. Trials, 17, 397 10.1186/s13063-016-1512-0 - DOI - PMC - PubMed
-
- Black, A. E. (2000). Critical evaluation of energy intake using the Goldberg cut‐off for energy intake: Basal metabolic rate. A practical guide to its calculation, use and limitations. International Journal of Obesity, 24, 1119–1130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

