The Unique Antimicrobial Recognition and Signaling Pathways in Tardigrades with a Comparison Across Ecdysozoa
- PMID: 31969428
- PMCID: PMC7056985
- DOI: 10.1534/g3.119.400734
The Unique Antimicrobial Recognition and Signaling Pathways in Tardigrades with a Comparison Across Ecdysozoa
Erratum in
-
Corrigendum.G3 (Bethesda). 2020 Jun 1;10(6):2127. doi: 10.1534/g3.120.401321. G3 (Bethesda). 2020. PMID: 32482730 Free PMC article. No abstract available.
Abstract
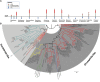
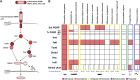
Tardigrades are microscopic animals known to withstand unfavorable abiotic conditions. These animals are also constantly exposed to biotic stresses, including parasites and internal microbiomes. However, the tardigrade immune mechanisms against these biotic stresses are largely uncharacterized. Due to the contentious phylogenetic position of tardigrades, it is not intuitive whether they possess an immune system more similar to that of arthropods (e.g., Toll, Imd, and JNK pathways of the Drosophila melanogaster antimicrobial response) or to that of nematodes (e.g., the Tir-1/Nsy-1/Sek-1/Pmk-1/Atf-7 signaling cassette [called Tir-1 pathway here]) in Caenorhabditis elegans). In this study, comparative genomic analyses were conducted to mine homologs of canonical D. melanogaster and C. elegans immune pathway genes from eight tardigrades (Echiniscoides cf. sigismundi, Echiniscus testudo, Hypsibius exemplaris, Mesobiotus philippinicus, Milnesium tardigradum, Paramacrobiotus richtersi, Richtersius cf. coronifer, and Ramazzottius varieornatus) and four non-arthropod ecdysozoans (two onychophorans: Epiperipatus sp. and Opisthopatus kwazululandi; one nematomorph: Paragordius varius; and one priapulan: Priapulus caudatus) in order to provide insights into the tardigrade antimicrobial system. No homologs of the intracellular components of the Toll pathway were detected in any of the tardigrades examined. Likewise, no homologs of most of the Imd pathway genes were detected in any of the tardigrades or any of the other non-arthropod ecdysozoans. Both the JNK and Tir-1 pathways, on the other hand, were found to be conserved across ecdysozoans. Interestingly, tardigrades had no detectable homologs of NF-κB, the major activator of antimicrobial response gene expression. Instead, tardigrades appear to possess NF-κB distantly related NFAT homologs. Overall, our results show that tardigrades have a unique gene pathway repertoire that differs from that of other ecdysozoans. Our study also provides a framework for future studies on tardigrade immune responses.
Keywords: Genetics of Immunity; biotic stress response; ecdysozoan immune response; tardigrade immune response.
Copyright © 2020 Mapalo et al.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
