Chromosome-Level Assembly of Drosophila bifasciata Reveals Important Karyotypic Transition of the X Chromosome
- PMID: 31969429
- PMCID: PMC7056972
- DOI: 10.1534/g3.119.400922
Chromosome-Level Assembly of Drosophila bifasciata Reveals Important Karyotypic Transition of the X Chromosome
Abstract
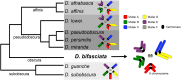
The Drosophila obscura species group is one of the most studied clades of Drosophila and harbors multiple distinct karyotypes. Here we present a de novo genome assembly and annotation of D. bifasciata, a species which represents an important subgroup for which no high-quality chromosome-level genome assembly currently exists. We combined long-read sequencing (Nanopore) and Hi-C scaffolding to achieve a highly contiguous genome assembly approximately 193 Mb in size, with repetitive elements constituting 30.1% of the total length. Drosophila bifasciata harbors four large metacentric chromosomes and the small dot, and our assembly contains each chromosome in a single scaffold, including the highly repetitive pericentromeres, which were largely composed of Jockey and Gypsy transposable elements. We annotated a total of 12,821 protein-coding genes and comparisons of synteny with D. athabasca orthologs show that the large metacentric pericentromeric regions of multiple chromosomes are conserved between these species. Importantly, Muller A (X chromosome) was found to be metacentric in D. bifasciata and the pericentromeric region appears homologous to the pericentromeric region of the fused Muller A-AD (XL and XR) of pseudoobscura/affinis subgroup species. Our finding suggests a metacentric ancestral X fused to a telocentric Muller D and created the large neo-X (Muller A-AD) chromosome ∼15 MYA. We also confirm the fusion of Muller C and D in D. bifasciata and show that it likely involved a centromere-centromere fusion.
Keywords: Muller element; Nanopore; centromere; chromosome.
Copyright © 2020 Bracewell et al.
Figures



References
-
- Buzzati-Traverso A. A., and Scossiroli R. E., 1955. The “Obscura Group” of the Genus Drosophila, pp. 47–92 in Advances in Genetics, edited by Demerec M., Academic Press, Cambridge, MA. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
