P3N-PIPO Interacts with P3 via the Shared N-Terminal Domain To Recruit Viral Replication Vesicles for Cell-to-Cell Movement
- PMID: 31969439
- PMCID: PMC7108826
- DOI: 10.1128/JVI.01898-19
P3N-PIPO Interacts with P3 via the Shared N-Terminal Domain To Recruit Viral Replication Vesicles for Cell-to-Cell Movement
Abstract
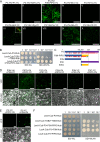
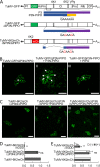
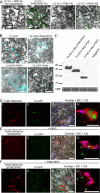
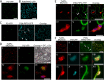
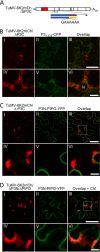
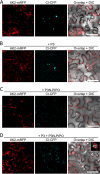
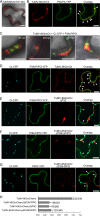
P3N-PIPO, the only dedicated movement protein (MP) of potyviruses, directs cylindrical inclusion (CI) protein from the cytoplasm to the plasmodesma (PD), where CI forms conical structures for intercellular movement. To better understand potyviral cell-to-cell movement, we further characterized P3N-PIPO using Turnip mosaic virus (TuMV) as a model virus. We found that P3N-PIPO interacts with P3 via the shared P3N domain and that TuMV mutants lacking the P3N domain of either P3N-PIPO or P3 are defective in cell-to-cell movement. Moreover, we found that the PIPO domain of P3N-PIPO is sufficient to direct CI to the PD, whereas the P3N domain is necessary for localization of P3N-PIPO to 6K2-labeled vesicles or aggregates. Finally, we discovered that the interaction between P3 and P3N-PIPO is essential for the recruitment of CI to cytoplasmic 6K2-containing structures and the association of 6K2-containing structures with PD-located CI inclusions. These data suggest that both P3N and PIPO domains are indispensable for potyviral cell-to-cell movement and that the 6K2 vesicles in proximity to PDs resulting from multipartite interactions among 6K2, P3, P3N-PIPO, and CI may also play an essential role in this process.IMPORTANCE Potyviruses include numerous economically important viruses that represent approximately 30% of known plant viruses. However, there is still limited information about the mechanism of potyviral cell-to-cell movement. Here, we show that P3N-PIPO interacts with and recruits CI to the PD via the PIPO domain and interacts with P3 via the shared P3N domain. We further report that the interaction of P3N-PIPO and P3 is associated with 6K2 vesicles and brings the 6K2 vesicles into proximity with PD-located CI structures. These results support the notion that the replication and cell-to-cell movement of potyviruses are processes coupled by anchoring viral replication complexes at the entrance of PDs, which greatly increase our knowledge of the intercellular movement of potyviruses.
Keywords: 6K2; CI; P3; P3N-PIPO; cell-to-cell movement; potyvirus; replication.
Copyright © 2020 American Society for Microbiology.
Figures

Similar articles
-
Cylindrical Inclusion Protein of Turnip Mosaic Virus Serves as a Docking Point for the Intercellular Movement of Viral Replication Vesicles.Plant Physiol. 2017 Dec;175(4):1732-1744. doi: 10.1104/pp.17.01484. Epub 2017 Oct 31. Plant Physiol. 2017. PMID: 29089395 Free PMC article.
-
Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement.PLoS Pathog. 2012;8(4):e1002639. doi: 10.1371/journal.ppat.1002639. Epub 2012 Apr 12. PLoS Pathog. 2012. PMID: 22511869 Free PMC article.
-
The C-terminal region of the Turnip mosaic virus P3 protein is essential for viral infection via targeting P3 to the viral replication complex.Virology. 2017 Oct;510:147-155. doi: 10.1016/j.virol.2017.07.016. Epub 2017 Jul 20. Virology. 2017. PMID: 28735115
-
Cell-to-cell movement of plant viruses via plasmodesmata: a current perspective on potyviruses.Curr Opin Virol. 2021 Jun;48:10-16. doi: 10.1016/j.coviro.2021.03.002. Epub 2021 Mar 27. Curr Opin Virol. 2021. PMID: 33784579 Review.
-
The mystery remains: How do potyviruses move within and between cells?Mol Plant Pathol. 2023 Dec;24(12):1560-1574. doi: 10.1111/mpp.13383. Epub 2023 Aug 12. Mol Plant Pathol. 2023. PMID: 37571979 Free PMC article. Review.
Cited by
-
The conserved aromatic residue W122 is a determinant of potyviral coat protein stability, replication, and cell-to-cell movement in plants.Mol Plant Pathol. 2021 Feb;22(2):189-203. doi: 10.1111/mpp.13017. Epub 2020 Nov 27. Mol Plant Pathol. 2021. PMID: 33245804 Free PMC article.
-
The 6-kilodalton peptide 1 in plant viruses of the family Potyviridae is a viroporin.Proc Natl Acad Sci U S A. 2024 May 21;121(21):e2401748121. doi: 10.1073/pnas.2401748121. Epub 2024 May 13. Proc Natl Acad Sci U S A. 2024. PMID: 38739789 Free PMC article.
-
A Genetic Study of Spillovers in the Bean Common Mosaic Subgroup of Potyviruses.Viruses. 2024 Aug 23;16(9):1351. doi: 10.3390/v16091351. Viruses. 2024. PMID: 39339828 Free PMC article.
-
Activation of MAPK-mediated immunity by phosphatidic acid in response to positive-strand RNA viruses.Plant Commun. 2024 Jan 8;5(1):100659. doi: 10.1016/j.xplc.2023.100659. Epub 2023 Jul 10. Plant Commun. 2024. PMID: 37434356 Free PMC article.
-
A Bifunctional Nuclease Promotes the Infection of Zucchini Yellow Mosaic Virus in Watermelon by Targeting P3.Plants (Basel). 2024 Dec 6;13(23):3431. doi: 10.3390/plants13233431. Plants (Basel). 2024. PMID: 39683224 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

