Regulation and Maturation of the Shewanella oneidensis Sulfite Reductase SirA
- PMID: 31969587
- PMCID: PMC6976685
- DOI: 10.1038/s41598-020-57587-6
Regulation and Maturation of the Shewanella oneidensis Sulfite Reductase SirA
Abstract
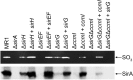
Shewanella oneidensis, a metal reducer and facultative anaerobe, expresses a large number of c-type cytochromes, many of which function as anaerobic reductases. All of these proteins contain the typical heme-binding motif CXXCH and require the Ccm proteins for maturation. Two c-type cytochrome reductases also possess atypical heme-binding sites, the NrfA nitrite reductase (CXXCK) and the SirA sulfite reductase (CX12NKGCH). S. oneidensis MR-1 encodes two cytochrome c synthetases (CcmF and SirE) and two apocytochrome c chaperones (CcmI and SirG). SirE located in the sir gene cluster is required for the maturation of SirA, but not NrfA. Here we show that maturation of SirA requires the combined function of the two apocytochrome c chaperones CcmI and SirG. Loss of either protein resulted in decreased sulfite reductase. Furthermore, SirA was not detected in a mutant that lacked both chaperones, perhaps due to misfolding or instability. These results suggest that CcmI interacts with SirEFG during SirA maturation, and with CcmF during maturation of NrfA. Additionally, we show that CRP regulates expression of sirA via the newly identified transcriptional regulatory protein, SirR.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

