Robust and persistent reactivation of SIV and HIV by N-803 and depletion of CD8+ cells
- PMID: 31969705
- PMCID: PMC7580846
- DOI: 10.1038/s41586-020-1946-0
Robust and persistent reactivation of SIV and HIV by N-803 and depletion of CD8+ cells
Erratum in
-
Author Correction: Robust and persistent reactivation of SIV and HIV by N-803 and depletion of CD8+ cells.Nature. 2020 Feb;578(7796):E21. doi: 10.1038/s41586-020-2002-9. Nature. 2020. PMID: 32015546
Abstract
Human immunodeficiency virus (HIV) persists indefinitely in individuals with HIV who receive antiretroviral therapy (ART) owing to a reservoir of latently infected cells that contain replication-competent virus1-4. Here, to better understand the mechanisms responsible for latency persistence and reversal, we used the interleukin-15 superagonist N-803 in conjunction with the depletion of CD8+ lymphocytes in ART-treated macaques infected with simian immunodeficiency virus (SIV). Although N-803 alone did not reactivate virus production, its administration after the depletion of CD8+ lymphocytes in conjunction with ART treatment induced robust and persistent reactivation of the virus in vivo. We found viraemia of more than 60 copies per ml in all macaques (n = 14; 100%) and in 41 out of a total of 56 samples (73.2%) that were collected each week after N-803 administration. Notably, concordant results were obtained in ART-treated HIV-infected humanized mice. In addition, we observed that co-culture with CD8+ T cells blocked the in vitro latency-reversing effect of N-803 on primary human CD4+ T cells that were latently infected with HIV. These results advance our understanding of the mechanisms responsible for latency reversal and lentivirus reactivation during ART-suppressed infection.
Conflict of interest statement
The authors of this paper declare no competing interests.
Figures

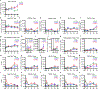





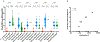



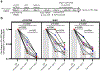

Comment in
-
Shocking HIV out of hiding.Nat Rev Immunol. 2020 Mar;20(3):138-139. doi: 10.1038/s41577-020-0283-8. Nat Rev Immunol. 2020. PMID: 32015471 No abstract available.
-
Reactivation of latent HIV moves shock-and-kill treatments forward.Nature. 2020 Feb;578(7793):42-43. doi: 10.1038/d41586-020-00010-x. Nature. 2020. PMID: 32020104 No abstract available.
-
Shocking HIV out of hiding.Nat Rev Drug Discov. 2020 Mar;19(3):167. doi: 10.1038/d41573-020-00023-1. Nat Rev Drug Discov. 2020. PMID: 32127671 No abstract available.
References
-
- UNAIDS. UNAIDS Data 2017. Joint United Nations; Program on HIV/AIDS (2017).
-
- Finzi D et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). - PubMed
-
- Wong JK et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH108179/MH/NIMH NIH HHS/United States
- 75N91019D00024/CA/NCI NIH HHS/United States
- UM1 AI126619/AI/NIAID NIH HHS/United States
- R01 AI143414/AI/NIAID NIH HHS/United States
- S10 OD025002/OD/NIH HHS/United States
- R01 AI125064/AI/NIAID NIH HHS/United States
- S10 OD026799/OD/NIH HHS/United States
- R01 AI111899/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- U42 OD011023/OD/NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- P51 OD011092/OD/NIH HHS/United States
- P30 AI050409/AI/NIAID NIH HHS/United States
- R01 AI123010/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- R01 AI090797/AI/NIAID NIH HHS/United States
- UM1 AI124436/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

