Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo
- PMID: 31969707
- PMCID: PMC7111210
- DOI: 10.1038/s41586-020-1951-3
Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo
Abstract
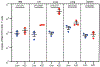
Long-lasting, latently infected resting CD4+ T cells are the greatest obstacle to obtaining a cure for HIV infection, as these cells can persist despite decades of treatment with antiretroviral therapy (ART). Estimates indicate that more than 70 years of continuous, fully suppressive ART are needed to eliminate the HIV reservoir1. Alternatively, induction of HIV from its latent state could accelerate the decrease in the reservoir, thus reducing the time to eradication. Previous attempts to reactivate latent HIV in preclinical animal models and in clinical trials have measured HIV induction in the peripheral blood with minimal focus on tissue reservoirs and have had limited effect2-9. Here we show that activation of the non-canonical NF-κB signalling pathway by AZD5582 results in the induction of HIV and SIV RNA expression in the blood and tissues of ART-suppressed bone-marrow-liver-thymus (BLT) humanized mice and rhesus macaques infected with HIV and SIV, respectively. Analysis of resting CD4+ T cells from tissues after AZD5582 treatment revealed increased SIV RNA expression in the lymph nodes of macaques and robust induction of HIV in almost all tissues analysed in humanized mice, including the lymph nodes, thymus, bone marrow, liver and lung. This promising approach to latency reversal-in combination with appropriate tools for systemic clearance of persistent HIV infection-greatly increases opportunities for HIV eradication.
Figures


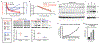








Comment in
-
Shocking HIV out of hiding.Nat Rev Immunol. 2020 Mar;20(3):138-139. doi: 10.1038/s41577-020-0283-8. Nat Rev Immunol. 2020. PMID: 32015471 No abstract available.
-
Reactivation of latent HIV moves shock-and-kill treatments forward.Nature. 2020 Feb;578(7793):42-43. doi: 10.1038/d41586-020-00010-x. Nature. 2020. PMID: 32020104 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 AI050409/AI/NIAID NIH HHS/United States
- R01 MH108179/MH/NIMH NIH HHS/United States
- R34 MH119963/MH/NIMH NIH HHS/United States
- UM1 AI126619/AI/NIAID NIH HHS/United States
- AI111899/AI/NIAID NIH HHS/United States
- AI123010/AI/NIAID NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- R56 AI117851/AI/NIAID NIH HHS/United States
- AI1117851/AI/NIAID NIH HHS/United States
- K24 AI143471/AI/NIAID NIH HHS/United States
- U19 AI096113/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- R01 AI123010/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- D43 TW009532/TW/FIC NIH HHS/United States
- S10 OD026799/OD/NIH HHS/United States
- R01 AI111899/AI/NIAID NIH HHS/United States
- 5P30CA016086-41/CA/NCI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- AI096113/AI/NIAID NIH HHS/United States
- MH108179/MH/NIMH NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- UM1 AI124436/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

