Targeting of temperate phages drives loss of type I CRISPR-Cas systems
- PMID: 31969710
- PMCID: PMC7007301
- DOI: 10.1038/s41586-020-1936-2
Targeting of temperate phages drives loss of type I CRISPR-Cas systems
Erratum in
-
Publisher Correction: Targeting of temperate phages drives loss of type I CRISPR-Cas systems.Nature. 2020 Mar;579(7799):E10. doi: 10.1038/s41586-020-2089-z. Nature. 2020. PMID: 32123354
Abstract
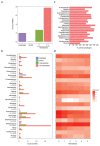
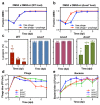
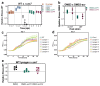
On infection of their host, temperate viruses that infect bacteria (bacteriophages; hereafter referred to as phages) enter either a lytic or a lysogenic cycle. The former results in lysis of bacterial cells and phage release (resulting in horizontal transmission), whereas lysogeny is characterized by the integration of the phage into the host genome, and dormancy (resulting in vertical transmission)1. Previous co-culture experiments using bacteria and mutants of temperate phages that are locked in the lytic cycle have shown that CRISPR-Cas systems can efficiently eliminate the invading phages2,3. Here we show that, when challenged with wild-type temperate phages (which can become lysogenic), type I CRISPR-Cas immune systems cannot eliminate the phages from the bacterial population. Furthermore, our data suggest that, in this context, CRISPR-Cas immune systems are maladaptive to the host, owing to the severe immunopathological effects that are brought about by imperfect matching of spacers to the integrated phage sequences (prophages). These fitness costs drive the loss of CRISPR-Cas from bacterial populations, unless the phage carries anti-CRISPR (acr) genes that suppress the immune system of the host. Using bioinformatics, we show that this imperfect targeting is likely to occur frequently in nature. These findings help to explain the patchy distribution of CRISPR-Cas immune systems within and between bacterial species, and highlight the strong selective benefits of phage-encoded acr genes for both the phage and the host under these circumstances.
Conflict of interest statement
The authors declare no competing interests.
Figures
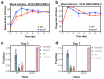

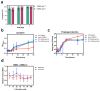

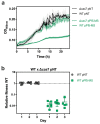
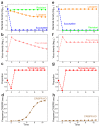




Similar articles
-
Bacteria exploit viral dormancy to establish CRISPR-Cas immunity.Cell Host Microbe. 2025 Mar 12;33(3):330-340.e6. doi: 10.1016/j.chom.2025.02.001. Epub 2025 Feb 25. Cell Host Microbe. 2025. PMID: 40010333
-
Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting.Nature. 2014 Oct 30;514(7524):633-7. doi: 10.1038/nature13637. Epub 2014 Aug 31. Nature. 2014. PMID: 25174707 Free PMC article.
-
Incomplete prophage tolerance by type III-A CRISPR-Cas systems reduces the fitness of lysogenic hosts.Nat Commun. 2018 Jan 4;9(1):61. doi: 10.1038/s41467-017-02557-2. Nat Commun. 2018. PMID: 29302058 Free PMC article.
-
Anti-CRISPRs go viral: The infection biology of CRISPR-Cas inhibitors.Cell Host Microbe. 2021 May 12;29(5):704-714. doi: 10.1016/j.chom.2020.12.007. Epub 2021 Jan 13. Cell Host Microbe. 2021. PMID: 33444542 Free PMC article. Review.
-
Lysogeny in the oceans: Lessons from cultivated model systems and a reanalysis of its prevalence.Environ Microbiol. 2020 Dec;22(12):4919-4933. doi: 10.1111/1462-2920.15233. Epub 2020 Sep 29. Environ Microbiol. 2020. PMID: 32935433 Review.
Cited by
-
Phages and their potential to modulate the microbiome and immunity.Cell Mol Immunol. 2021 Apr;18(4):889-904. doi: 10.1038/s41423-020-00532-4. Epub 2020 Sep 8. Cell Mol Immunol. 2021. PMID: 32901128 Free PMC article. Review.
-
CRISPR-Cas Controls Cryptic Prophages.Int J Mol Sci. 2022 Dec 19;23(24):16195. doi: 10.3390/ijms232416195. Int J Mol Sci. 2022. PMID: 36555835 Free PMC article.
-
Transient eco-evolutionary dynamics early in a phage epidemic have strong and lasting impact on the long-term evolution of bacterial defences.PLoS Biol. 2023 Sep 15;21(9):e3002122. doi: 10.1371/journal.pbio.3002122. eCollection 2023 Sep. PLoS Biol. 2023. PMID: 37713428 Free PMC article.
-
Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements.Nat Commun. 2020 Nov 6;11(1):5652. doi: 10.1038/s41467-020-19415-3. Nat Commun. 2020. PMID: 33159058 Free PMC article.
-
Adaptation by Type V-A and V-B CRISPR-Cas Systems Demonstrates Conserved Protospacer Selection Mechanisms Between Diverse CRISPR-Cas Types.CRISPR J. 2022 Aug;5(4):536-547. doi: 10.1089/crispr.2021.0150. Epub 2022 Jul 12. CRISPR J. 2022. PMID: 35833800 Free PMC article.
References
-
- Stewart FM, Levin BR. The population biology of bacterial viruses: Why be temperate. Theor Popul Biol. 1984;26:93–117. - PubMed
-
- Westra ER, et al. Parasite Exposure Drives Selective Evolution of Constitutive versus Inducible Defense. Curr Biol. 2015;25:1043–1049. - PubMed
-
- Barrangou R, et al. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science. 2017;315:1709–1712. - PubMed
-
- Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

