Proneural and mesenchymal glioma stem cells display major differences in splicing and lncRNA profiles
- PMID: 31969990
- PMCID: PMC6965107
- DOI: 10.1038/s41525-019-0108-5
Proneural and mesenchymal glioma stem cells display major differences in splicing and lncRNA profiles
Abstract
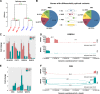
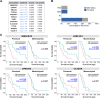
Therapy resistance and recurrence in high-grade gliomas are driven by their populations of glioma stem cells (GSCs). Thus, detailed molecular characterization of GSCs is needed to develop more effective therapies. We conducted a study to identify differences in the splicing profile and expression of long non-coding RNAs in proneural and mesenchymal GSC cell lines. Genes related to cell cycle, DNA repair, cilium assembly, and splicing showed the most differences between GSC subgroups. We also identified genes distinctly associated with survival among patients of mesenchymal or proneural subgroups. We determined that multiple long non-coding RNAs with increased expression in mesenchymal GSCs are associated with poor survival of glioblastoma patients. In summary, our study established critical differences between proneural and mesenchymal GSCs in splicing profiles and expression of long non-coding RNA. These splicing isoforms and lncRNA signatures may contribute to the uniqueness of GSC subgroups, thus contributing to cancer phenotypes and explaining differences in therapeutic responses.
Keywords: Cancer genomics; Cancer stem cells; Gene expression analysis; Gene regulatory networks.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials

