Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption
- PMID: 31970406
- PMCID: PMC7102970
- DOI: 10.1093/nar/gkaa010
Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption
Abstract
During oocyte maturation, changes in gene expression depend exclusively on translation and degradation of maternal mRNAs rather than transcription. Execution of this translation program is essential for assembling the molecular machinery required for meiotic progression, fertilization, and embryo development. With the present study, we used a RiboTag/RNA-Seq approach to explore the timing of maternal mRNA translation in quiescent oocytes as well as in oocytes progressing through the first meiotic division. This genome-wide analysis reveals a global switch in maternal mRNA translation coinciding with oocyte re-entry into the meiotic cell cycle. Messenger RNAs whose translation is highly active in quiescent oocytes invariably become repressed during meiotic re-entry, whereas transcripts repressed in quiescent oocytes become activated. Experimentally, we have defined the exact timing of the switch and the repressive function of CPE elements, and identified a novel role for CPEB1 in maintaining constitutive translation of a large group of maternal mRNAs during maturation.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


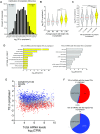
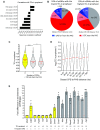
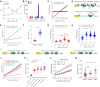
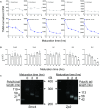


References
-
- Klemm S.L., Shipony Z., Greenleaf W.J.. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019; 20:207–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

