Genetic structure and admixture in sheep from terminal breeds in the United States
- PMID: 31970815
- PMCID: PMC7065203
- DOI: 10.1111/age.12905
Genetic structure and admixture in sheep from terminal breeds in the United States
Abstract
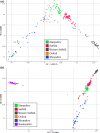
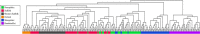
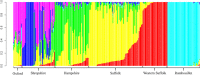
Selection for performance in diverse production settings has resulted in variation across sheep breeds worldwide. Although sheep are an important species to the United States, the current genetic relationship among many terminal sire breeds is not well characterized. Suffolk, Hampshire, Shropshire and Oxford (terminal) and Rambouillet (dual purpose) sheep (n = 248) sampled from different flocks were genotyped using the Applied Biosystems Axiom Ovine Genotyping Array (50K), and additional Shropshire sheep (n = 26) using the Illumina Ovine SNP50 BeadChip. Relationships were investigated by calculating observed heterozygosity, inbreeding coefficients, eigenvalues, pairwise Wright's FST estimates and an identity by state matrix. The mean observed heterozygosity for each breed ranged from 0.30 to 0.35 and was consistent with data reported in other US and Australian sheep. Suffolk from two different regions of the United States (Midwest and West) clustered separately in eigenvalue plots and the rectangular cladogram. Further, divergence was detected between Suffolk from different regions with Wright's FST estimate. Shropshire animals showed the greatest divergence from other terminal breeds in this study. Admixture between breeds was examined using admixture, and based on cross-validation estimates, the best fit number of populations (clusters) was K = 6. The greatest admixture was observed within Hampshire, Suffolk, and Shropshire breeds. When plotting eigenvalues, US terminal breeds clustered separately in comparison with sheep from other locations of the world. Understanding the genetic relationships between terminal sire breeds in sheep will inform us about the potential applicability of markers derived in one breed to other breeds based on relatedness.
Keywords: genetic admixture; genetic relationships; sheep; terminal sheep breeds.
© 2020 The Authors. Animal Genetics published by John Wiley & Sons Ltd on behalf of Stichting International Foundation for Animal Genetics.
Figures

References
-
- American Sheep Industry (2011) National Animal Health Monitoring System (NAHMS) Sheep 2011 Study. https://sheepusa.org/researcheducation-animalhealth-nahms.
-
- Andersson L. (2012) How selective sweeps in domesticated animals provide new insight into biological mechanisms. Journal of Internal Medicine 271, 1–14. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

