Confinement and Crowding Effects on Folding of a Multidomain Y-Family DNA Polymerase
- PMID: 31972079
- PMCID: PMC7258223
- DOI: 10.1021/acs.jctc.9b01146
Confinement and Crowding Effects on Folding of a Multidomain Y-Family DNA Polymerase
Abstract
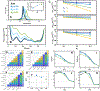
Proteins in vivo endure highly various interactions from the luxuriant surrounding macromolecular cosolutes. Confinement and macromolecular crowding are the two major effects that should be considered while comparing the results of protein dynamics from in vitro to in vivo. However, efforts have been largely focused on single domain protein folding up to now, and the quantifications of the in vivo effects in terms of confinements and crowders on modulating the structure and dynamics as well as the physical understanding of the underlying mechanisms on multidomain protein folding are still challenging. Here we developed a topology-based model to investigate folding of a multidomain Y-family DNA polymerase (DPO4) within spherical confined space and in the presence of repulsive and attractive crowders. We uncovered that the entropic component of the thermodynamic driving force led by confinements and repulsive crowders increases the stability of folded states relative to the folding intermediates and unfolded states, while the enthalpic component of the thermodynamic driving force led by attractive crowders gives rise to the opposite effects with less stability. We found that the shapes of DPO4 conformations influenced by the confinements and the crowders are quite different even when only the entropic component of the thermodynamic driving force is considered. We uncovered that under all in vivo conditions, the folding cooperativity of DPO4 decreases compared to that in bulk. We showed that the loss of folding cooperativity can promote the sequential domain-wise folding, which was widely found in cotranslational multidomain protein folding, and effectively prohibit the backtracking led by topological frustrations during multidomain protein folding processes.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
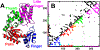



References
-
- Fulton AB How crowded is the cytoplasm? Cell 1982, 30, 345–347. - PubMed
-
- Ellis RJ; Minton AP Cell biology: join the crowd. Nature 2003, 425, 27. - PubMed
-
- Zimmerman SB; Minton AP Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct 1993, 22, 27–65. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
