Association Between Sulfur-Metabolizing Bacterial Communities in Stool and Risk of Distal Colorectal Cancer in Men
- PMID: 31972239
- PMCID: PMC7384232
- DOI: 10.1053/j.gastro.2019.12.029
Association Between Sulfur-Metabolizing Bacterial Communities in Stool and Risk of Distal Colorectal Cancer in Men
Abstract
Background & aims: Sulfur-metabolizing microbes, which convert dietary sources of sulfur into genotoxic hydrogen sulfide (H2S), have been associated with development of colorectal cancer (CRC). We identified a dietary pattern associated with sulfur-metabolizing bacteria in stool and then investigated its association with risk of incident CRC using data from a large prospective study of men.
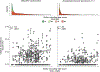
Methods: We collected data from 51,529 men enrolled in the Health Professionals Follow-up Study since 1986 to determine the association between sulfur-metabolizing bacteria in stool and risk of CRC over 26 years of follow-up. First, in a subcohort of 307 healthy men, we profiled serial stool metagenomes and metatranscriptomes and assessed diet using semiquantitative food frequency questionnaires to identify food groups associated with 43 bacterial species involved in sulfur metabolism. We used these data to develop a sulfur microbial dietary score. We then used Cox proportional hazards modeling to evaluate adherence to this pattern among eligible individuals (n = 48,246) from 1986 through 2012 with risk for incident CRC.
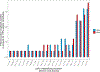
Results: Foods associated with higher sulfur microbial diet scores included increased consumption of processed meats and low-calorie drinks and lower consumption of vegetables and legumes. Increased sulfur microbial diet scores were associated with risk of distal colon and rectal cancers, after adjusting for other risk factors (multivariable relative risk, highest vs lowest quartile, 1.43; 95% confidence interval 1.14-1.81; P-trend = .002). In contrast, sulfur microbial diet scores were not associated with risk of proximal colon cancer (multivariable relative risk 0.86; 95% CI 0.65-1.14; P-trend = .31).
Conclusions: In an analysis of participants in the Health Professionals Follow-up Study, we found that long-term adherence to a dietary pattern associated with sulfur-metabolizing bacteria in stool was associated with an increased risk of distal CRC. Further studies are needed to determine how sulfur-metabolizing bacteria might contribute to CRC pathogenesis.
Keywords: Cancer Biogeography; Colorectal Carcinogenesis; FFQ; Fecal Microbes.
Copyright © 2020 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures
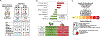


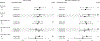
Similar articles
-
Association Between the Sulfur Microbial Diet and Risk of Colorectal Cancer.JAMA Netw Open. 2021 Nov 1;4(11):e2134308. doi: 10.1001/jamanetworkopen.2021.34308. JAMA Netw Open. 2021. PMID: 34767023 Free PMC article.
-
The Sulfur Microbial Diet Is Associated With Increased Risk of Early-Onset Colorectal Cancer Precursors.Gastroenterology. 2021 Nov;161(5):1423-1432.e4. doi: 10.1053/j.gastro.2021.07.008. Epub 2021 Jul 14. Gastroenterology. 2021. PMID: 34273347 Free PMC article.
-
The Sulfur Microbial Diet and Risk of Colorectal Cancer by Molecular Subtypes and Intratumoral Microbial Species in Adult Men.Clin Transl Gastroenterol. 2021 Aug 1;12(8):e00338. doi: 10.14309/ctg.0000000000000338. Clin Transl Gastroenterol. 2021. PMID: 34333506 Free PMC article.
-
Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer.Gastroenterology. 2020 Jan;158(2):322-340. doi: 10.1053/j.gastro.2019.06.048. Epub 2019 Oct 3. Gastroenterology. 2020. PMID: 31586566 Free PMC article. Review.
-
Dietary Patterns and Breast, Colorectal, Lung, and Prostate Cancer: A Systematic Review [Internet].Alexandria (VA): USDA Nutrition Evidence Systematic Review; 2020 Jul. Alexandria (VA): USDA Nutrition Evidence Systematic Review; 2020 Jul. PMID: 35129907 Free Books & Documents. Review.
Cited by
-
Reproducible and opposing gut microbiome signatures distinguish autoimmune diseases and cancers: a systematic review and meta-analysis.Microbiome. 2022 Dec 9;10(1):218. doi: 10.1186/s40168-022-01373-1. Microbiome. 2022. PMID: 36482486 Free PMC article.
-
Implications of hydrogen sulfide in colorectal cancer: Mechanistic insights and diagnostic and therapeutic strategies.Redox Biol. 2023 Feb;59:102601. doi: 10.1016/j.redox.2023.102601. Epub 2023 Jan 7. Redox Biol. 2023. PMID: 36630819 Free PMC article. Review.
-
Processed Food and Atopic Dermatitis: A Pooled Analysis of Three Cross-Sectional Studies in Chinese Adults.Front Nutr. 2021 Dec 6;8:754663. doi: 10.3389/fnut.2021.754663. eCollection 2021. Front Nutr. 2021. PMID: 34938758 Free PMC article.
-
Associations between the Gut Microbiota, Race, and Ethnicity of Patients with Colorectal Cancer: A Pilot and Feasibility Study.Cancers (Basel). 2023 Sep 13;15(18):4546. doi: 10.3390/cancers15184546. Cancers (Basel). 2023. PMID: 37760515 Free PMC article.
-
Meta-Analysis of Altered Gut Microbiota Reveals Microbial and Metabolic Biomarkers for Colorectal Cancer.Microbiol Spectr. 2022 Aug 31;10(4):e0001322. doi: 10.1128/spectrum.00013-22. Epub 2022 Jun 29. Microbiol Spectr. 2022. PMID: 35766483 Free PMC article.
References
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. - PubMed
-
- Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 2000;343:78–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- C10674/A27140/CRUK_/Cancer Research UK/United Kingdom
- P30 DK043351/DK/NIDDK NIH HHS/United States
- U01 CA152904/CA/NCI NIH HHS/United States
- K24 DK098311/DK/NIDDK NIH HHS/United States
- 27140/CRUK_/Cancer Research UK/United Kingdom
- K07 CA218377/CA/NCI NIH HHS/United States
- R00 CA215314/CA/NCI NIH HHS/United States
- K01 DK120742/DK/NIDDK NIH HHS/United States
- L30 DK118604/DK/NIDDK NIH HHS/United States
- K99 DK119412/DK/NIDDK NIH HHS/United States
- R35 CA197735/CA/NCI NIH HHS/United States
- K01 DK110267/DK/NIDDK NIH HHS/United States
- R01 CA202704/CA/NCI NIH HHS/United States
- P01 CA055075/CA/NCI NIH HHS/United States
- R01 CA151993/CA/NCI NIH HHS/United States
- U01 CA167552/CA/NCI NIH HHS/United States
- L30 CA209764/CA/NCI NIH HHS/United States
- K23 DK125838/DK/NIDDK NIH HHS/United States
- U54 DE023798/DE/NIDCR NIH HHS/United States
- T32 CA009001/CA/NCI NIH HHS/United States
- UM1 CA167552/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

