Characterization of the Rumen Microbiota and Volatile Fatty Acid Profiles of Weaned Goat Kids under Shrub-Grassland Grazing and Indoor Feeding
- PMID: 31972989
- PMCID: PMC7070841
- DOI: 10.3390/ani10020176
Characterization of the Rumen Microbiota and Volatile Fatty Acid Profiles of Weaned Goat Kids under Shrub-Grassland Grazing and Indoor Feeding
Abstract
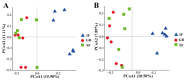
In this study, we conducted comparative analyses to characterize the rumen microbiota and volatile fatty acid (VFA) profiles of weaned Nanjiang Yellow goat kids under shrub-grassland grazing (GR), shrub-grassland grazing and supplementary feeding (SF), and indoor feeding (IF) systems. We observed significant differences (p < 0.05) in the concentrations of total VFA and the proportions of acetate and butyrate in the rumen fluid among the three groups, whereas the proportions of propionate and the acetate/propionate ratio did not differ substantially. Alpha diversity of the rumen bacterial and archaeal populations in the GR and SF kids was significantly higher (p < 0.05) than that in the IF goat kids, and significant differences (p < 0.05) in similarity were observed in the comparisons of GR vs. IF and SF vs. IF. The most predominant bacterial phyla were Bacteroidetes and Firmicutes across the three groups, and the archaeal community was mainly composed of Euryarchaeota. At the genus and species levels, the cellulose-degrading bacteria, including Lachnospiraceae, Ruminococcaceae and Butyrivibrio fibrisolvens, were abundant in the GR and SF groups. Furthermore, 27 bacterial and 11 unique archaeal taxa, such as Lachnospiraceae, Butyrivibrio fibrisolvens, and Methanobrevibacter ruminantium, were identified as biomarkers, and showed significantly different (p < 0.05) abundances among the three groups. Significant Spearman correlations (p < 0.05), between the abundances of several microbial biomarkers and the concentrations of VFAs, were further observed. In summary, our results demonstrated that the adaptation to grazing required more rumen bacterial populations due to complex forage types in shrub-grassland, although the rumen fermentation pattern did not change substantially among the three feeding systems. Some microbial taxa could be used as biomarkers for different feeding systems, particularly cellulose-degrading bacteria associated with grazing.
Keywords: 16S rRNA gene; goat; grazing; microbiota; rumen; volatile fatty acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Distinct Stage Changes in Early-Life Colonization and Acquisition of the Gut Microbiota and Its Correlations With Volatile Fatty Acids in Goat Kids.Front Microbiol. 2020 Oct 9;11:584742. doi: 10.3389/fmicb.2020.584742. eCollection 2020. Front Microbiol. 2020. PMID: 33162961 Free PMC article.
-
Shrub coverage alters the rumen bacterial community of yaks (Bos grunniens) grazing in alpine meadows.J Anim Sci Technol. 2020 Jul;62(4):504-520. doi: 10.5187/jast.2020.62.4.504. Epub 2020 Jul 31. J Anim Sci Technol. 2020. PMID: 32803183 Free PMC article.
-
Shift of Feeding Strategies from Grazing to Different Forage Feeds Reshapes the Rumen Microbiota To Improve the Ability of Tibetan Sheep (Ovis aries) To Adapt to the Cold Season.Microbiol Spectr. 2023 Feb 27;11(2):e0281622. doi: 10.1128/spectrum.02816-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36809032 Free PMC article.
-
A Comparison of Growth Performance, Blood Parameters, Rumen Fermentation, and Bacterial Community of Tibetan Sheep When Fattened by Pasture Grazing versus Stall Feeding.Microorganisms. 2024 Sep 28;12(10):1967. doi: 10.3390/microorganisms12101967. Microorganisms. 2024. PMID: 39458276 Free PMC article.
-
Effects of supplementary feeding on the rumen morphology and bacterial diversity in lambs.PeerJ. 2020 Jun 19;8:e9353. doi: 10.7717/peerj.9353. eCollection 2020. PeerJ. 2020. PMID: 32596052 Free PMC article.
Cited by
-
Microbiota in Goat Buck Ejaculates Differs Between Breeding and Non-breeding Seasons.Front Vet Sci. 2022 May 13;9:867671. doi: 10.3389/fvets.2022.867671. eCollection 2022. Front Vet Sci. 2022. PMID: 35647092 Free PMC article.
-
Distinct Stage Changes in Early-Life Colonization and Acquisition of the Gut Microbiota and Its Correlations With Volatile Fatty Acids in Goat Kids.Front Microbiol. 2020 Oct 9;11:584742. doi: 10.3389/fmicb.2020.584742. eCollection 2020. Front Microbiol. 2020. PMID: 33162961 Free PMC article.
-
Effects of Grazing in a Low Deciduous Forest on Rumen Microbiota and Volatile Fatty Acid Production in Lambs.Animals (Basel). 2025 May 27;15(11):1565. doi: 10.3390/ani15111565. Animals (Basel). 2025. PMID: 40509031 Free PMC article.
-
Physical, Metabolic, and Microbial Rumen Development in Goat Kids: A Review on the Challenges and Strategies of Early Weaning.Animals (Basel). 2023 Jul 26;13(15):2420. doi: 10.3390/ani13152420. Animals (Basel). 2023. PMID: 37570229 Free PMC article. Review.
-
The gas production, ruminal fermentation parameters, and microbiota in response to Clostridium butyricum supplementation on in vitro varying with media pH levels.Front Microbiol. 2022 Sep 21;13:960623. doi: 10.3389/fmicb.2022.960623. eCollection 2022. Front Microbiol. 2022. PMID: 36212861 Free PMC article.
References
-
- Wang Y., McAllister T.A. Rumen Microbes, Enzymes and Feed Digestion-A Review. Asian Australas J. Anim. Sci. 2002;15:1659–1676. doi: 10.5713/ajas.2002.1659. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

