Quantification of Endogenous Brain Tissue Displacement Imaging by Radiofrequency Ultrasound
- PMID: 31973031
- PMCID: PMC7168898
- DOI: 10.3390/diagnostics10020057
Quantification of Endogenous Brain Tissue Displacement Imaging by Radiofrequency Ultrasound
Abstract
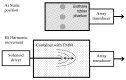
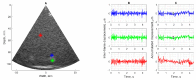
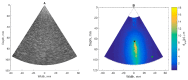
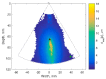
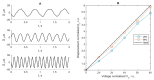
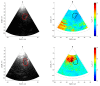
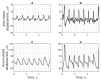
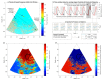
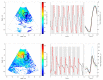
The purpose of this paper is a quantification of displacement parameters used in the imaging of brain tissue endogenous motion using ultrasonic radiofrequency (RF) signals. In a preclinical study, an ultrasonic diagnostic system with RF output was equipped with dedicated signal processing software and subject head-ultrasonic transducer stabilization. This allowed the use of RF scanning frames for the calculation of micrometer-range displacements, excluding sonographer-induced motions. Analysis of quantitative displacement estimates in dynamical phantom experiments showed that displacements of 55 µm down to 2 µm were quantified as confident according to Pearson correlation between signal fragments (minimum p ≤ 0.001). The same algorithm and scanning hardware were used in experiments and clinical imaging which allows translating phantom results to Alzheimer's disease patients and healthy elderly subjects as examples. The confident quantitative displacement waveforms of six in vivo heart-cycle episodes ranged from 8 µm up to 263 µm (Pearson correlation p ≤ 0.01). Displacement time sequences showed promising possibilities to evaluate the morphology of endogenous displacement signals at each point of the scanning plane, while displacement maps-regional distribution of displacement parameters-were essential for tissue characterization.
Keywords: brain; radiofrequency ultrasound; tissue displacements; transcranial sonography.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hachemi M.E., Remeniéras J., Desmidt T., Camus V., Tranquart F. Elderly depression diagnostic of diabetic patients by brain tissue pulsatility imaging. Phys. Proc. 2010;3:713–718. doi: 10.1016/j.phpro.2010.01.090. - DOI
-
- Shiina T., Nightingale K.R., Palmeri M.L., Hall T.J., Bamber J.C., Barr R.G., Castera L., Choi B.I., Chou Y.-H., Cosgrove D., et al. WFUMB guidelines and recommendations for clinical use of ultrasound elastography: Part 1: Basic principles and terminology. Ultrasound Med Biol. 2015;41:1126–1147. doi: 10.1016/j.ultrasmedbio.2015.03.009. - DOI - PubMed
LinkOut - more resources
Full Text Sources

