The Flagellar Gene Regulates Biofilm Formation and Mussel Larval Settlement and Metamorphosis
- PMID: 31973189
- PMCID: PMC7036800
- DOI: 10.3390/ijms21030710
The Flagellar Gene Regulates Biofilm Formation and Mussel Larval Settlement and Metamorphosis
Abstract
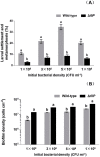
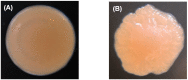
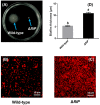
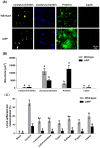
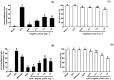
Biofilms are critical components of most marine systems and provide biochemical cues that can significantly impact overall community composition. Although progress has been made in the bacteria-animal interaction, the molecular basis of modulation of settlement and metamorphosis in most marine animals by bacteria is poorly understood. Here, Pseudoalteromonas marina showing inducing activity on mussel settlement and metamorphosis was chosen as a model to clarify the mechanism that regulates the bacteria-mussel interaction. We constructed a flagellin synthetic protein gene fliP deletion mutant of P. marina and checked whether deficiency of fliP gene will impact inducing activity, motility, and extracellular polymeric substances of biofilms. Furthermore, we examined the effect of flagellar proteins extracted from bacteria on larval settlement and metamorphosis. The deletion of the fliP gene caused the loss of the flagella structure and motility of the ∆fliP strain. Deficiency of the fliP gene promoted the biofilm formation and changed biofilm matrix by reducing β-polysaccharides and increasing extracellular proteins and finally reduced biofilm-inducing activities. Flagellar protein extract promoted mussel metamorphosis, and ∆fliP biofilms combined with additional flagellar proteins induced similar settlement and metamorphosis rate compared to that of the wild-type strain. These findings provide novel insight on the molecular interactions between bacteria and mussels.
Keywords: Mytilus coruscus; biofilm; flagellar gene; larval settlement and metamorphosis; mussel.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Chemotaxis gene of a bacterium impacts larval settlement and metamorphosis in the marine mussel Mytilus coruscus via c-di-GMP controlling extracellular protein production.Biofouling. 2024 Nov;40(10):882-892. doi: 10.1080/08927014.2024.2423806. Epub 2024 Nov 14. Biofouling. 2024. PMID: 39540578
-
A bacterial polysaccharide biosynthesis-related gene inversely regulates larval settlement and metamorphosis of Mytilus coruscus.Biofouling. 2020 Aug;36(7):753-765. doi: 10.1080/08927014.2020.1807520. Epub 2020 Aug 26. Biofouling. 2020. PMID: 32847400
-
Monospecific Biofilms of Pseudoalteromonas Promote Larval Settlement and Metamorphosis of Mytilus coruscus.Sci Rep. 2020 Feb 13;10(1):2577. doi: 10.1038/s41598-020-59506-1. Sci Rep. 2020. PMID: 32054934 Free PMC article.
-
Love at First Taste: Induction of Larval Settlement by Marine Microbes.Int J Mol Sci. 2020 Jan 22;21(3):731. doi: 10.3390/ijms21030731. Int J Mol Sci. 2020. PMID: 31979128 Free PMC article. Review.
-
Biofouling and antifouling.Nat Prod Rep. 2004 Feb;21(1):94-104. doi: 10.1039/b302231p. Epub 2003 Nov 25. Nat Prod Rep. 2004. PMID: 15039837 Review.
Cited by
-
Chemical signaling in biofilm-mediated biofouling.Nat Chem Biol. 2024 Nov;20(11):1406-1419. doi: 10.1038/s41589-024-01740-z. Epub 2024 Sep 30. Nat Chem Biol. 2024. PMID: 39349970 Review.
-
Deep-sea bacteria trigger settlement and metamorphosis of the mussel Mytilus coruscus larvae.Sci Rep. 2021 Jan 13;11(1):919. doi: 10.1038/s41598-020-79832-8. Sci Rep. 2021. PMID: 33441694 Free PMC article.
-
Anti-Biofilm Effects of Z102-E of Lactiplantibacillus plantarum against Listeria monocytogenes and the Mechanism Revealed by Transcriptomic Analysis.Foods. 2024 Aug 8;13(16):2495. doi: 10.3390/foods13162495. Foods. 2024. PMID: 39200422 Free PMC article.
-
Chemically mediated rheotaxis of endangered tri-spine horseshoe crab: potential dispersing mechanism to vegetated nursery habitats along the coast.PeerJ. 2022 Dec 5;10:e14465. doi: 10.7717/peerj.14465. eCollection 2022. PeerJ. 2022. PMID: 36523452 Free PMC article.
-
Bacterial c-di-GMP triggers metamorphosis of mussel larvae through a STING receptor.NPJ Biofilms Microbiomes. 2024 Jun 20;10(1):51. doi: 10.1038/s41522-024-00523-7. NPJ Biofilms Microbiomes. 2024. PMID: 38902226 Free PMC article.
References
-
- Dobretsov S. Inhibition and induction of marine biofouling by biofilms. In: Flemming H.C., Murthy P.S., Venkatesan R., Cooksey K., editors. Marine And Industrial Biofouling. Springer; Berlin/Heidelberg, Germany: 2009. pp. 293–313.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

