Lung Microbiota Predict Clinical Outcomes in Critically Ill Patients
- PMID: 31973575
- PMCID: PMC7047465
- DOI: 10.1164/rccm.201907-1487OC
Lung Microbiota Predict Clinical Outcomes in Critically Ill Patients
Abstract
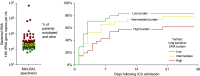
Rationale: Recent studies have revealed that, in critically ill patients, lung microbiota are altered and correlate with alveolar inflammation. The clinical significance of altered lung bacteria in critical illness is unknown.Objectives: To determine if clinical outcomes of critically ill patients are predicted by features of the lung microbiome at the time of admission.Methods: We performed a prospective, observational cohort study in an ICU at a university hospital. Lung microbiota were quantified and characterized using droplet digital PCR and bacterial 16S ribosomal RNA gene quantification and sequencing. Primary predictors were the bacterial burden, community diversity, and community composition of lung microbiota. The primary outcome was ventilator-free days, determined at 28 days after admission.Measurements and Main Results: Lungs of 91 critically ill patients were sampled using miniature BAL within 24 hours of ICU admission. Patients with increased lung bacterial burden had fewer ventilator-free days (hazard ratio, 0.43; 95% confidence interval, 0.21-0.88), which remained significant when the analysis was controlled for pneumonia and severity of illness. The community composition of lung bacteria predicted ventilator-free days (P = 0.003), driven by the presence of gut-associated bacteria (e.g., species of the Lachnospiraceae and Enterobacteriaceae families). Detection of gut-associated bacteria was also associated with the presence of acute respiratory distress syndrome.Conclusions: Key features of the lung microbiome (bacterial burden and enrichment with gut-associated bacteria) predict outcomes in critically ill patients. The lung microbiome is an understudied source of clinical variation in critical illness and represents a novel therapeutic target for the prevention and treatment of acute respiratory failure.
Keywords: acute respiratory distress syndrome; lung injury; lung microbiome; prognosis.
Figures


Comment in
-
In the Wrong Place at the Wrong Time: Microbial Misplacement and Acute Respiratory Distress Syndrome.Am J Respir Crit Care Med. 2020 Mar 1;201(5):506-507. doi: 10.1164/rccm.202001-0004ED. Am J Respir Crit Care Med. 2020. PMID: 31973580 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

