Multi-trait Genomic Prediction Model Increased the Predictive Ability for Agronomic and Malting Quality Traits in Barley (Hordeum vulgare L.)
- PMID: 31974097
- PMCID: PMC7056970
- DOI: 10.1534/g3.119.400968
Multi-trait Genomic Prediction Model Increased the Predictive Ability for Agronomic and Malting Quality Traits in Barley (Hordeum vulgare L.)
Abstract
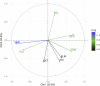
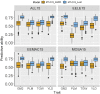
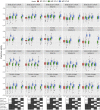
Plant breeders regularly evaluate multiple traits across multiple environments, which opens an avenue for using multiple traits in genomic prediction models. We assessed the potential of multi-trait (MT) genomic prediction model through evaluating several strategies of incorporating multiple traits (eight agronomic and malting quality traits) into the prediction models with two cross-validation schemes (CV1, predicting new lines with genotypic information only and CV2, predicting partially phenotyped lines using both genotypic and phenotypic information from correlated traits) in barley. The predictive ability was similar for single (ST-CV1) and multi-trait (MT-CV1) models to predict new lines. However, the predictive ability for agronomic traits was considerably increased when partially phenotyped lines (MT-CV2) were used. The predictive ability for grain yield using the MT-CV2 model with other agronomic traits resulted in 57% and 61% higher predictive ability than ST-CV1 and MT-CV1 models, respectively. Therefore, complex traits such as grain yield are better predicted when correlated traits are used. Similarly, a considerable increase in the predictive ability of malting quality traits was observed when correlated traits were used. The predictive ability for grain protein content using the MT-CV2 model with both agronomic and malting traits resulted in a 76% higher predictive ability than ST-CV1 and MT-CV1 models. Additionally, the higher predictive ability for new environments was obtained for all traits using the MT-CV2 model compared to the MT-CV1 model. This study showed the potential of improving the genomic prediction of complex traits by incorporating the information from multiple traits (cost-friendly and easy to measure traits) collected throughout breeding programs which could assist in speeding up breeding cycles.
Keywords: GenPred; Genomic Prediction; Multi-trait; Shared Data Resources; genomic prediction; grain yield, grain quality; malting quality; multi-environment.
Copyright © 2020 Bhatta et al.
Figures



References
-
- Bertholdsson N. O., 1999. Characterization of malting barley cultivars with more or less stable grain protein content under varying environmental conditions. Eur. J. Agron. 10: 1–8. 10.1016/S1161-0301(98)00043-4 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
