Pulse-chase SILAC-based analyses reveal selective oversynthesis and rapid turnover of mitochondrial protein components of respiratory complexes
- PMID: 31974161
- PMCID: PMC7049976
- DOI: 10.1074/jbc.RA119.011791
Pulse-chase SILAC-based analyses reveal selective oversynthesis and rapid turnover of mitochondrial protein components of respiratory complexes
Abstract
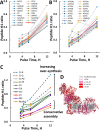
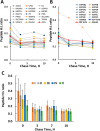
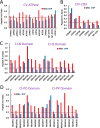
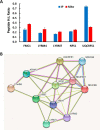
Mammalian mitochondria assemble four complexes of the respiratory chain (RCI, RCIII, RCIV, and RCV) by combining 13 polypeptides synthesized within mitochondria on mitochondrial ribosomes (mitoribosomes) with over 70 polypeptides encoded in nuclear DNA, translated on cytoplasmic ribosomes, and imported into mitochondria. We have previously observed that mitoribosome assembly is inefficient because some mitoribosomal proteins are produced in excess, but whether this is the case for other mitochondrial assemblies such as the RCs is unclear. We report here that pulse-chase stable isotope labeling with amino acids in cell culture (SILAC) is a valuable technique to study RC assembly because it can reveal considerable differences in the assembly rates and efficiencies of the different complexes. The SILAC analyses of HeLa cells indicated that assembly of RCV, comprising F1/Fo-ATPase, is rapid with little excess subunit synthesis, but that assembly of RCI (i.e. NADH dehydrogenase) is far less efficient, with dramatic oversynthesis of numerous proteins, particularly in the matrix-exposed N and Q domains. Unassembled subunits were generally degraded within 3 h. We also observed differential assembly kinetics for individual complexes that were immunoprecipitated with complex-specific antibodies. Immunoprecipitation with an antibody that recognizes the ND1 subunit of RCI co-precipitated a number of proteins implicated in FeS cluster assembly and newly synthesized ubiquinol-cytochrome c reductase Rieske iron-sulfur polypeptide 1 (UQCRFS1), the Rieske FeS protein in RCIII, reflecting some coordination between RCI and RCIII assemblies. We propose that pulse-chase SILAC labeling is a useful tool for studying rates of protein complex assembly and degradation.
Keywords: NADH dehydrogenase; mitochondria; mitochondrial biogenesis; mitochondrial respiratory chain complex; oxidative phosphorylation system; protein assembly; protein dynamics; protein synthesis; protein turnover; proteomics; respiratory complex assembly; stable isotope labeling with amino acids in cell culture (SILAC).
© 2020 Bogenhagen and Haley.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

