ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) releases latent defense signals in stems with reduced lignin content
- PMID: 31974310
- PMCID: PMC7022211
- DOI: 10.1073/pnas.1914422117
ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) releases latent defense signals in stems with reduced lignin content
Abstract
There is considerable interest in engineering plant cell wall components, particularly lignin, to improve forage quality and biomass properties for processing to fuels and bioproducts. However, modifying lignin content and/or composition in transgenic plants through down-regulation of lignin biosynthetic enzymes can induce expression of defense response genes in the absence of biotic or abiotic stress. Arabidopsis thaliana lines with altered lignin through down-regulation of hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT) or loss of function of cinnamoyl CoA reductase 1 (CCR1) express a suite of pathogenesis-related (PR) protein genes. The plants also exhibit extensive cell wall remodeling associated with induction of multiple cell wall-degrading enzymes, a process which renders the corresponding biomass a substrate for growth of the cellulolytic thermophile Caldicellulosiruptor bescii lacking a functional pectinase gene cluster. The cell wall remodeling also results in the release of size- and charge-heterogeneous pectic oligosaccharide elicitors of PR gene expression. Genetic analysis shows that both in planta PR gene expression and release of elicitors are the result of ectopic expression in xylem of the gene ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1), which is normally expressed during anther and silique dehiscence. These data highlight the importance of pectin in cell wall integrity and the value of lignin modification as a tool to interrogate the informational content of plant cell walls.
Keywords: cell wall remodeling; defense response; elicitor; lignin modification; polygalacturonase.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
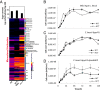
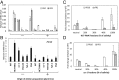



Comment in
-
Pectin-derived immune elicitors in response to lignin modification in plants.Proc Natl Acad Sci U S A. 2020 Mar 3;117(9):4442-4444. doi: 10.1073/pnas.2000509117. Epub 2020 Feb 6. Proc Natl Acad Sci U S A. 2020. PMID: 32029583 Free PMC article. No abstract available.
Similar articles
-
Activation tagging of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 promotes hypocotyl elongation, leaf expansion, stem lignification, mechanical stiffening, and lodging.Plant J. 2017 Mar;89(6):1159-1173. doi: 10.1111/tpj.13453. Epub 2017 Feb 11. Plant J. 2017. PMID: 28004869
-
ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis.Plant Cell. 2009 Jan;21(1):216-33. doi: 10.1105/tpc.108.063768. Epub 2009 Jan 23. Plant Cell. 2009. PMID: 19168715 Free PMC article.
-
FERONIA and wall-associated kinases coordinate defense induced by lignin modification in plant cell walls.Sci Adv. 2023 Mar 10;9(10):eadf7714. doi: 10.1126/sciadv.adf7714. Epub 2023 Mar 10. Sci Adv. 2023. PMID: 36897948 Free PMC article.
-
A review of xylan and lignin biosynthesis: foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes.Crit Rev Biochem Mol Biol. 2014 May-Jun;49(3):212-41. doi: 10.3109/10409238.2014.889651. Epub 2014 Feb 24. Crit Rev Biochem Mol Biol. 2014. PMID: 24564339 Review.
-
Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity.Phytochemistry. 2002 Oct;61(3):221-94. doi: 10.1016/s0031-9422(02)00211-x. Phytochemistry. 2002. PMID: 12359514 Review.
Cited by
-
Cell wall integrity regulation across plant species.Plant Mol Biol. 2022 Jul;109(4-5):483-504. doi: 10.1007/s11103-022-01284-7. Epub 2022 Jun 8. Plant Mol Biol. 2022. PMID: 35674976 Free PMC article. Review.
-
A primary cell wall cellulose-dependent defense mechanism against vascular pathogens revealed by time-resolved dual transcriptomics.BMC Biol. 2021 Aug 17;19(1):161. doi: 10.1186/s12915-021-01100-6. BMC Biol. 2021. PMID: 34404410 Free PMC article.
-
Post-Synthetic Reduction of Pectin Methylesterification Causes Morphological Abnormalities and Alterations to Stress Response in Arabidopsis thaliana.Plants (Basel). 2020 Nov 12;9(11):1558. doi: 10.3390/plants9111558. Plants (Basel). 2020. PMID: 33198397 Free PMC article.
-
Global transcriptional modulation and nutritional status of soybean plants following foliar application of zinc borate as a suspension concentrate fertilizer.Sci Rep. 2025 Jan 26;15(1):3309. doi: 10.1038/s41598-025-87771-5. Sci Rep. 2025. PMID: 39865117 Free PMC article.
-
Genomic imprinted genes in reciprocal hybrid endosperm of Brassica napus.BMC Plant Biol. 2021 Mar 16;21(1):140. doi: 10.1186/s12870-021-02908-8. BMC Plant Biol. 2021. PMID: 33726676 Free PMC article.
References
-
- Voxeur A., Höfte H., Cell wall integrity signaling in plants: “To grow or not to grow that’s the question.” Glycobiology 26, 950–960 (2016). - PubMed
-
- Chen F., Dixon R. A., Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761 (2007). - PubMed
-
- Shadle G., et al. , Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68, 1521–1529 (2007). - PubMed
-
- Kim J. I., Ciesielski P. N., Donohoe B. S., Chapple C., Li X., Chemically induced conditional rescue of the reduced epidermal fluorescence8 mutant of Arabidopsis reveals rapid restoration of growth and selective turnover of secondary metabolite pools. Plant Physiol. 164, 584–595 (2014). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

