The Medical Genome Reference Bank contains whole genome and phenotype data of 2570 healthy elderly
- PMID: 31974348
- PMCID: PMC6978518
- DOI: 10.1038/s41467-019-14079-0
The Medical Genome Reference Bank contains whole genome and phenotype data of 2570 healthy elderly
Abstract
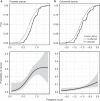
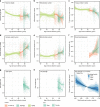
Population health research is increasingly focused on the genetic determinants of healthy ageing, but there is no public resource of whole genome sequences and phenotype data from healthy elderly individuals. Here we describe the first release of the Medical Genome Reference Bank (MGRB), comprising whole genome sequence and phenotype of 2570 elderly Australians depleted for cancer, cardiovascular disease, and dementia. We analyse the MGRB for single-nucleotide, indel and structural variation in the nuclear and mitochondrial genomes. MGRB individuals have fewer disease-associated common and rare germline variants, relative to both cancer cases and the gnomAD and UK Biobank cohorts, consistent with risk depletion. Age-related somatic changes are correlated with grip strength in men, suggesting blood-derived whole genomes may also provide a biologic measure of age-related functional deterioration. The MGRB provides a broadly applicable reference cohort for clinical genetics and genomic association studies, and for understanding the genetics of healthy ageing.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

