Sodium-induced population shift drives activation of thrombin
- PMID: 31974511
- PMCID: PMC6978324
- DOI: 10.1038/s41598-020-57822-0
Sodium-induced population shift drives activation of thrombin
Abstract
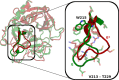
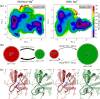
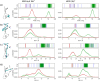
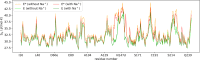
The equilibrium between active E and inactive E* forms of thrombin is assumed to be governed by the allosteric binding of a Na+ ion. Here we use molecular dynamics simulations and Markov state models to sample transitions between active and inactive states. With these calculations we are able to compare thermodynamic and kinetic properties depending on the presence of Na+. For the first time, we directly observe sodium-induced conformational changes in long-timescale computer simulations. Thereby, we are able to explain the resulting change in activity. We observe a stabilization of the active form in presence of Na+ and a shift towards the inactive form in Na+-free simulations. We identify key structural features to quantify and monitor this conformational shift. These include the accessibility of the S1 pocket and the reorientation of W215, of R221a and of the Na+ loop. The structural characteristics exhibit dynamics at various timescales: Conformational changes in the Na+ binding loop constitute the slowest observed movement. Depending on its orientation, it induces conformational shifts in the nearby substrate binding site. Only after this shift, residue W215 is able to move freely, allowing thrombin to adopt a binding-competent conformation.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Na+ Binding Is Ineffective in Forming a Primary Substrate Pocket of Thrombin.J Phys Chem B. 2016 Nov 23;120(46):11873-11879. doi: 10.1021/acs.jpcb.6b07827. Epub 2016 Nov 11. J Phys Chem B. 2016. PMID: 27781431
-
Expression of allosteric linkage between the sodium ion binding site and exosite I of thrombin during prothrombin activation.J Biol Chem. 2007 Jun 1;282(22):16095-104. doi: 10.1074/jbc.M610577200. Epub 2007 Apr 12. J Biol Chem. 2007. PMID: 17430903 Free PMC article.
-
Molecular dissection of Na+ binding to thrombin.J Biol Chem. 2004 Jul 23;279(30):31842-53. doi: 10.1074/jbc.M401756200. Epub 2004 May 19. J Biol Chem. 2004. PMID: 15152000
-
[Sodium ions as the effector of catalytic action of alpha-thrombin].Ukr Biokhim Zh (1999). 2007 Jan-Feb;79(1):5-21. Ukr Biokhim Zh (1999). 2007. PMID: 18030731 Review. Russian.
-
How Na+ activates thrombin--a review of the functional and structural data.Biol Chem. 2008 Aug;389(8):1025-35. doi: 10.1515/BC.2008.113. Biol Chem. 2008. PMID: 18979627 Free PMC article. Review.
Cited by
-
Shedding Light on the Molecular Recognition of Sub-Kilodalton Macrocyclic Peptides on Thrombin by Supervised Molecular Dynamics.Front Mol Biosci. 2021 Aug 31;8:707661. doi: 10.3389/fmolb.2021.707661. eCollection 2021. Front Mol Biosci. 2021. PMID: 34532343 Free PMC article.
-
Exosite Binding in Thrombin: A Global Structural/Dynamic Overview of Complexes with Aptamers and Other Ligands.Int J Mol Sci. 2021 Oct 6;22(19):10803. doi: 10.3390/ijms221910803. Int J Mol Sci. 2021. PMID: 34639143 Free PMC article. Review.
-
Relationship between serum sodium level and sepsis-induced coagulopathy.Front Med (Lausanne). 2024 Jan 8;10:1324369. doi: 10.3389/fmed.2023.1324369. eCollection 2023. Front Med (Lausanne). 2024. PMID: 38298508 Free PMC article.
-
Energy penalties enhance flexible receptor docking in a model cavity.Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2106195118. doi: 10.1073/pnas.2106195118. Proc Natl Acad Sci U S A. 2021. PMID: 34475217 Free PMC article.
-
19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.J Biol Chem. 2020 Jun 12;295(24):8227-8235. doi: 10.1074/jbc.RA120.013419. Epub 2020 May 1. J Biol Chem. 2020. PMID: 32358061 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

